Similar presentations:
Віруси з олДНК
1.
ВІРУСИ З олДНК2.
Стадії реплікації олДНК вірусів1. Перетворення в дволанцюжкову ДНК
клітинний процес відновлення?)
2. Рання транскрипція (клітинні
ферменти)
3. Трансляція (регуляторних) білків і
“кільцева" оцДНК реплікація
4. Пізня транскрипція (як правило,
вірус-опосередкованих білків)
5. Синтез пізніх (= структурних)
білків
6. "Ізоляція" вірусного генома
оцДНК
7. Збірка у віріони
3. ssDNA Viruses
Bacteria ("bacteriophages", eg. Inoviridae,Microviridae),
Mammals (Circoviridae, Parvoviridae)
Birds (circovirus-like organisms),
Plants (Geminiviridae, banana bunchy top-like viruses
or Nanoviruses).
4. Parvovirus
Family: Parvoviridae~20 nm in diameter
Latin parvus means small
(0.02 µm)
Single-stranded DNA virus
Icosahedral capsid
No envelope
X-ray crystallographic image of parvovirus
5.
Infection Cycle6. Genome Structure
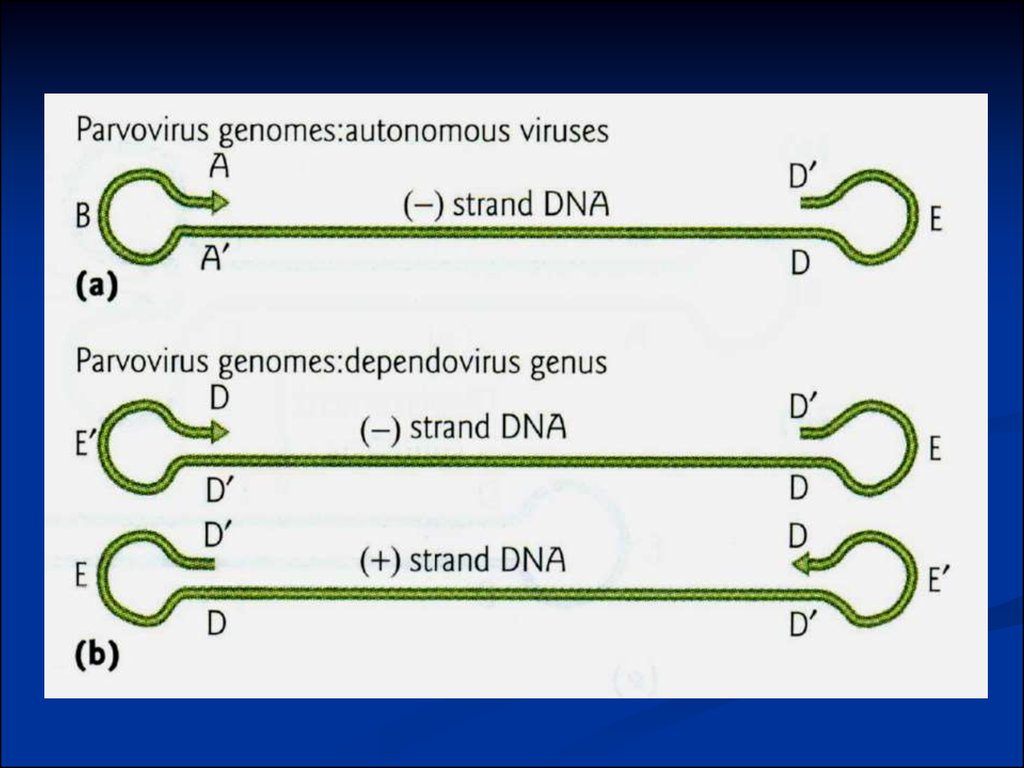
Type:Autonomous virus: (-) DNA
eg. Mice minute virus
Defective virus: (+) and (-) DNA in separate virions
Adeno-associated virus
Replication dependents on coinfection with helper virus.
Eg. Adenovirus
Terminal hairpins (inverted reapeats)
7.
8.
Adeno-associated virus (AAV); ssDNAITR
ITR
Rep
Cap
5 kb ssDNA, inverted terminal repeats (ITR)
Rep gene required for DNA replication
Cap gene encodes capsid proteins
9. Генетична карта парвовірусу (ААВ)
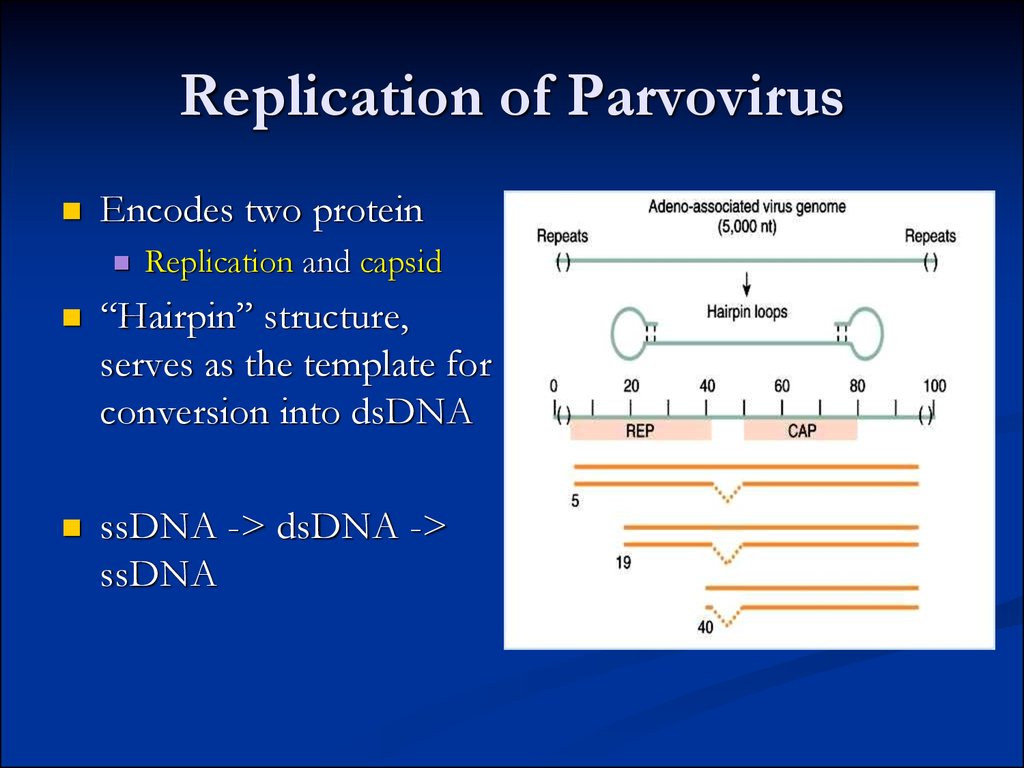
Valeriy Polischuk Virology Department Kyiv National Taras Shevchenko University10. Replication of Parvovirus
Encodes two proteinReplication and capsid
“Hairpin” structure,
serves as the template for
conversion into dsDNA
ssDNA -> dsDNA ->
ssDNA
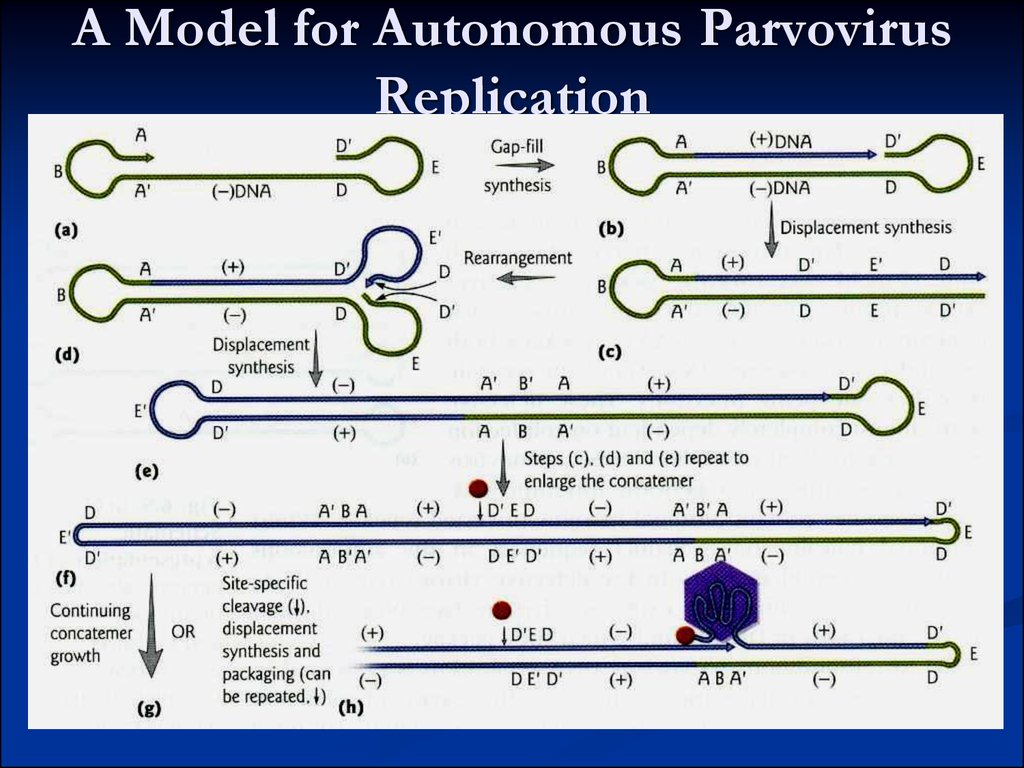
11. A Model for Autonomous Parvovirus Replication
12.
Autonomous parvovirus Life CycleFrom Medical Microbiology, 5th ed.,
Murray, Rosenthal, Kobayashi & Pfaller,
Mosby Inc., 2002, Fig. 56-2.
13.
Helper dependent parvovirus (AAV) replicationInfection with adenovirus
Infection without adenovirus
Lytic
replication
AAV DNA
integrates into
chromosome 19
Superinfect
with
adenovirus
14.
15.
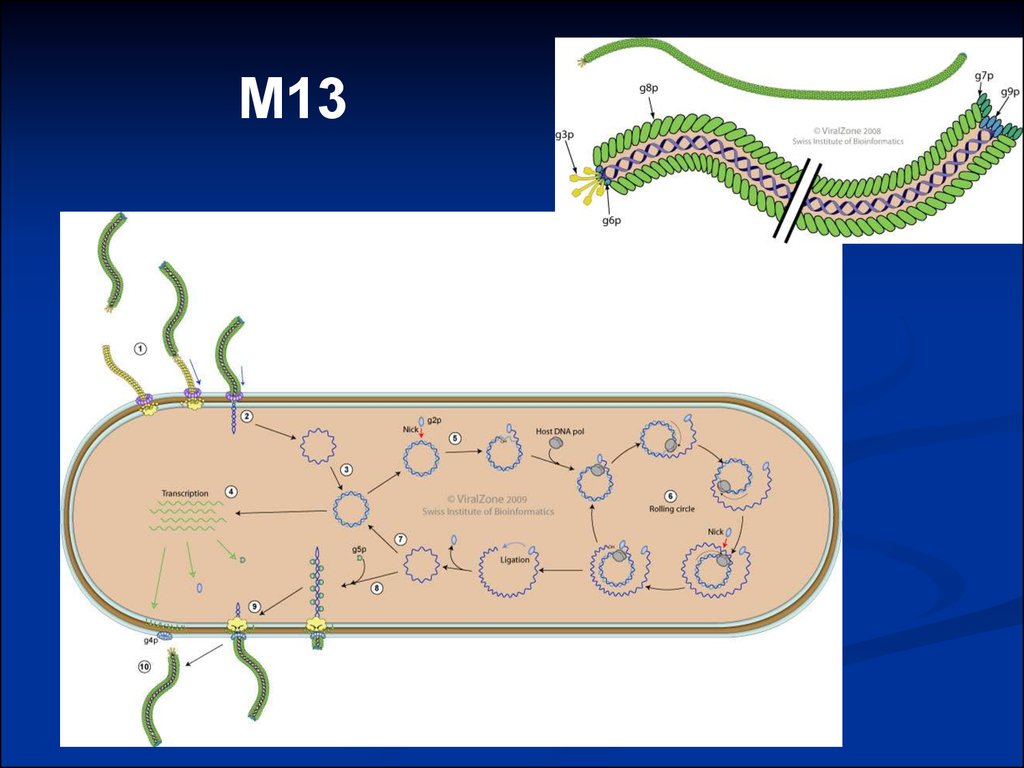
M1316.
Schematic representation of bacteriophageM13 replication cycle
E.coli
F-pilus
gp 8
DNA
gp 5
M13
17.
Schematic structure of bacteriophage M13 filamentDistal end
Gene 7 protein
Gene 9 protein
Circular ssDNA
Gene 8 protein
Gene 6 protein
Proximal end
Gene 3 protein
18. M13 Structure
Dimensions:6.5nm in diameter
Length dependant on genome but wild type
approximately 930nm
Mass 16.3MD of which 87% comprised of
protein
Genome:
Single stranded circular DNA molecule,
covalently closed
Housed in a flexible protein cylinder
19. M13 Structure
ProteinsLength of phage cylinder comprises 2700 copies of
the 50-amino-acid major coat protein pVIII
At one terminus:
5 copies of 33 AA residue pVII
5 copies of 32 AA residue pIX
At one terminus:
5 copies of 406 AA residue pIII
5 copies of 112 AA residue pVI
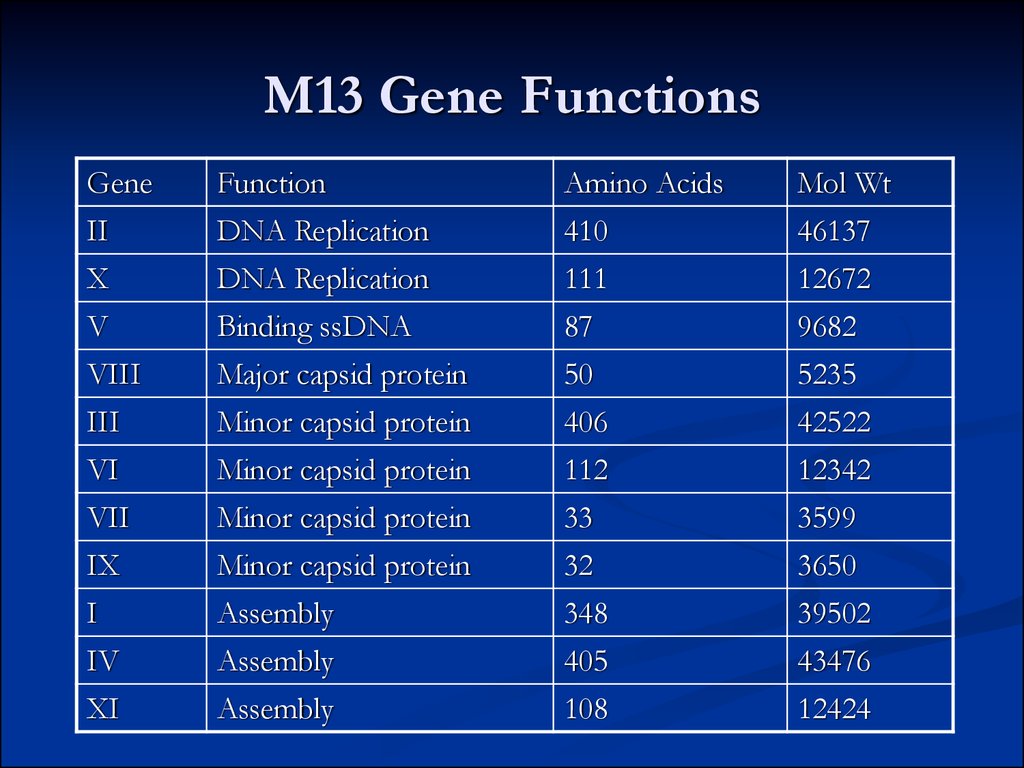
20. M13 Gene Functions
GeneFunction
Amino Acids
Mol Wt
II
DNA Replication
410
46137
X
DNA Replication
111
12672
V
Binding ssDNA
87
9682
VIII
Major capsid protein
50
5235
III
Minor capsid protein
406
42522
VI
Minor capsid protein
112
12342
VII
Minor capsid protein
33
3599
IX
Minor capsid protein
32
3650
I
Assembly
348
39502
IV
Assembly
405
43476
XI
Assembly
108
12424
21.
M13 (or f1) phages are filamentous phages that infect E. coli through pili,and are able to produce new virions without lysing the host cell. M13 has some key
structural elements:
Circular, single-stranded DNA
~6.4 kb long
10 genes in the genome.
22.
Of these 10 genes, we are concerned with the followingthree:
Gene II: codes for nickase, allows rolling-circle
replication
Gene III: codes for the pilot protein, which guides the
nascent ssDNA to the membrane,
Gene VIII: coat protein, encapsulates the pilot protein
and the ssDNA phage DNA as it extrudes through the
membrane
M13 phages are useful for a number of applications:
sequencing
mutagenesis
probes
lambda-ZAP subcloning
phage display libraries


















 biology
biology








