Similar presentations:
Mobile genetic elements
1. Last Class
DNA replication
Chromosome replication
DNA repair
General Recombination
2.
Site-specific recombinationMoves specialized nucleotide sequence (mobile
genetic elements) between non-homologous sites
within a genome.
Transpositional site-specific recombination
Conservative site-specific recombinatinon
3.
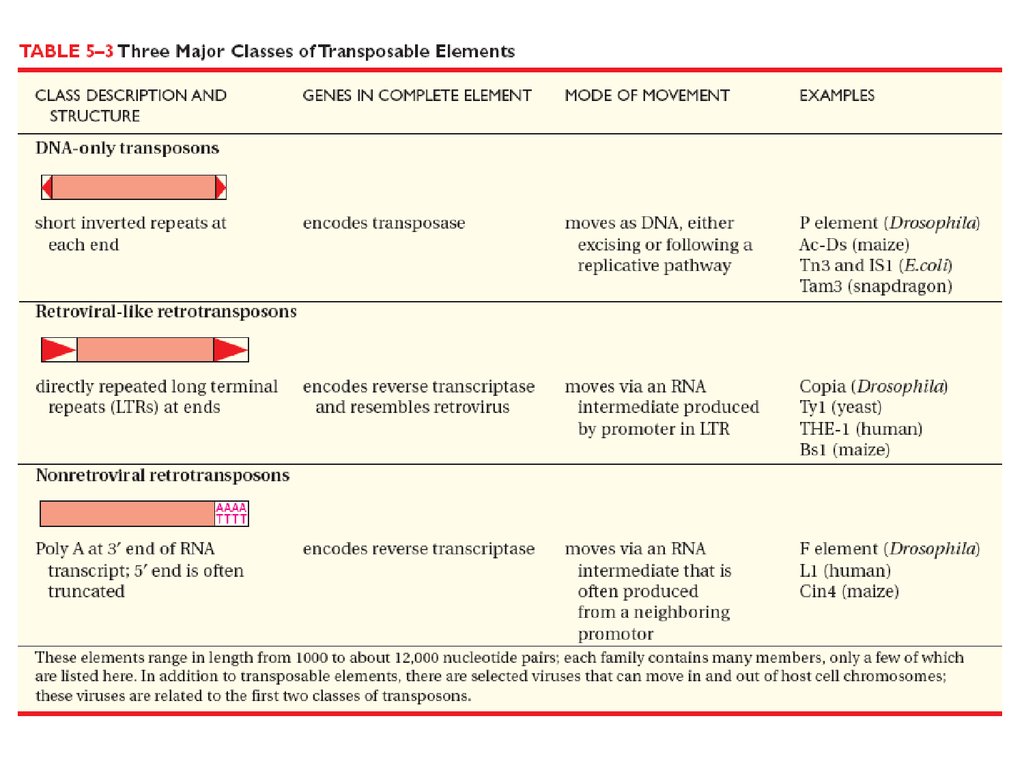
Transpositional site-specificrecombination
Modest target site selectivity and insert mobile
genetic elements into many sites
Transposase enzyme cuts out mobile genetic
elements and insert them into specific sites.
4.
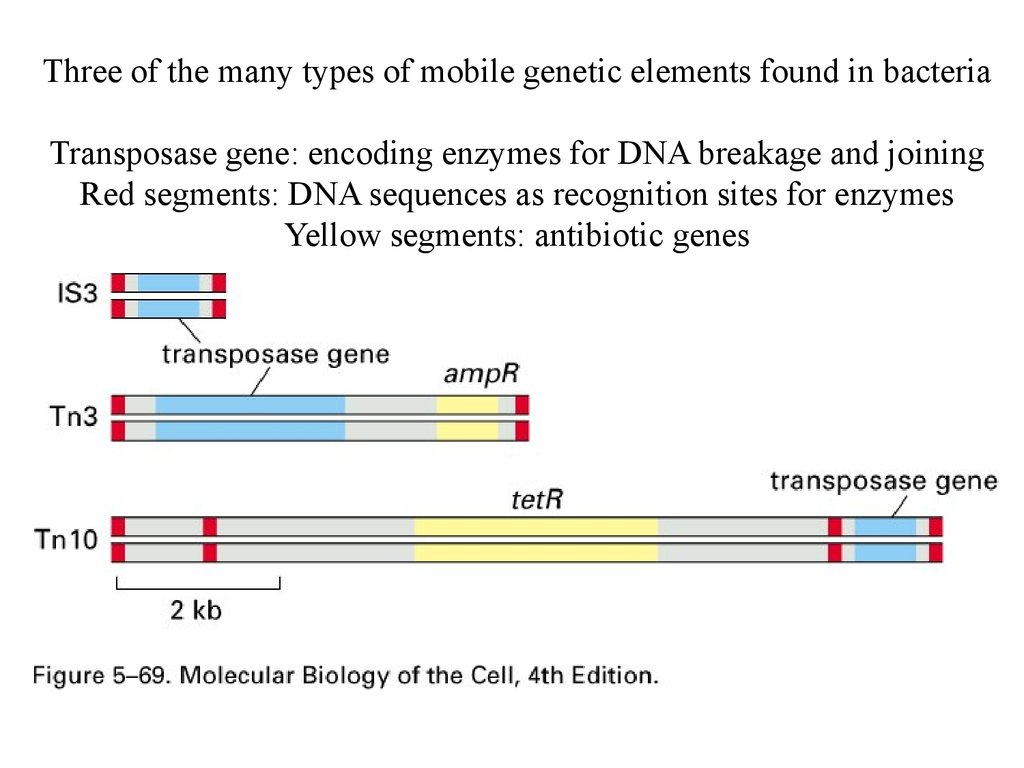
Three of the many types of mobile genetic elements found in bacteriaTransposase gene: encoding enzymes for DNA breakage and joining
Red segments: DNA sequences as recognition sites for enzymes
Yellow segments: antibiotic genes
5.
6.
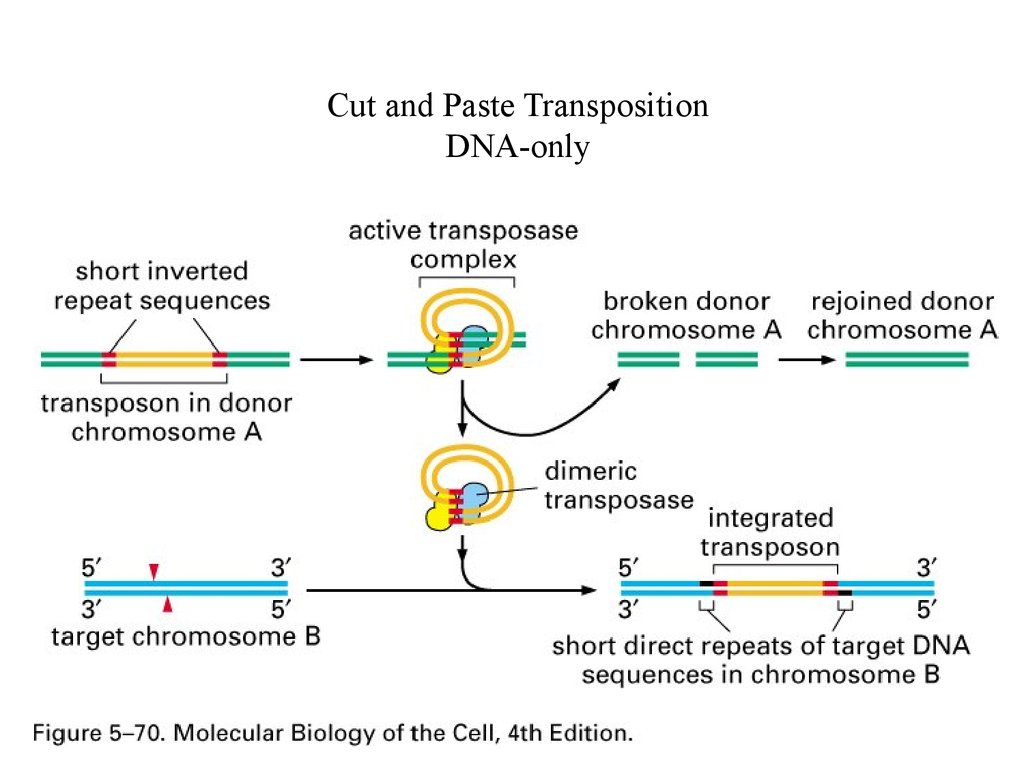
Cut and Paste TranspositionDNA-only
7.
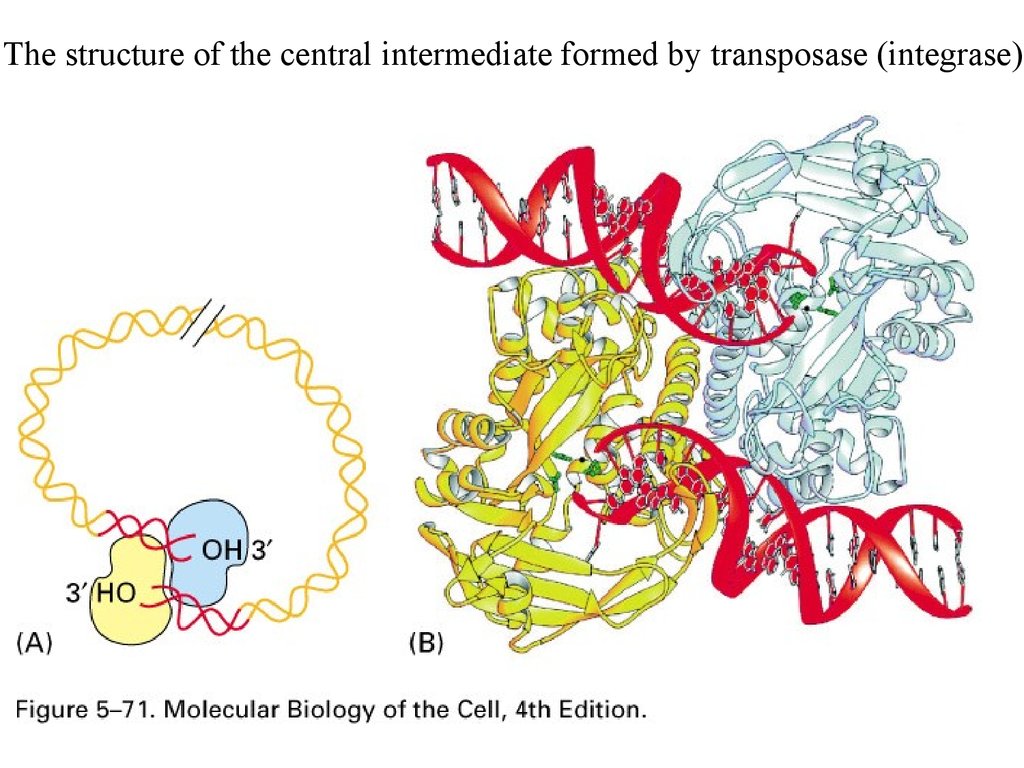
The structure of the central intermediate formed by transposase (integrase)8.
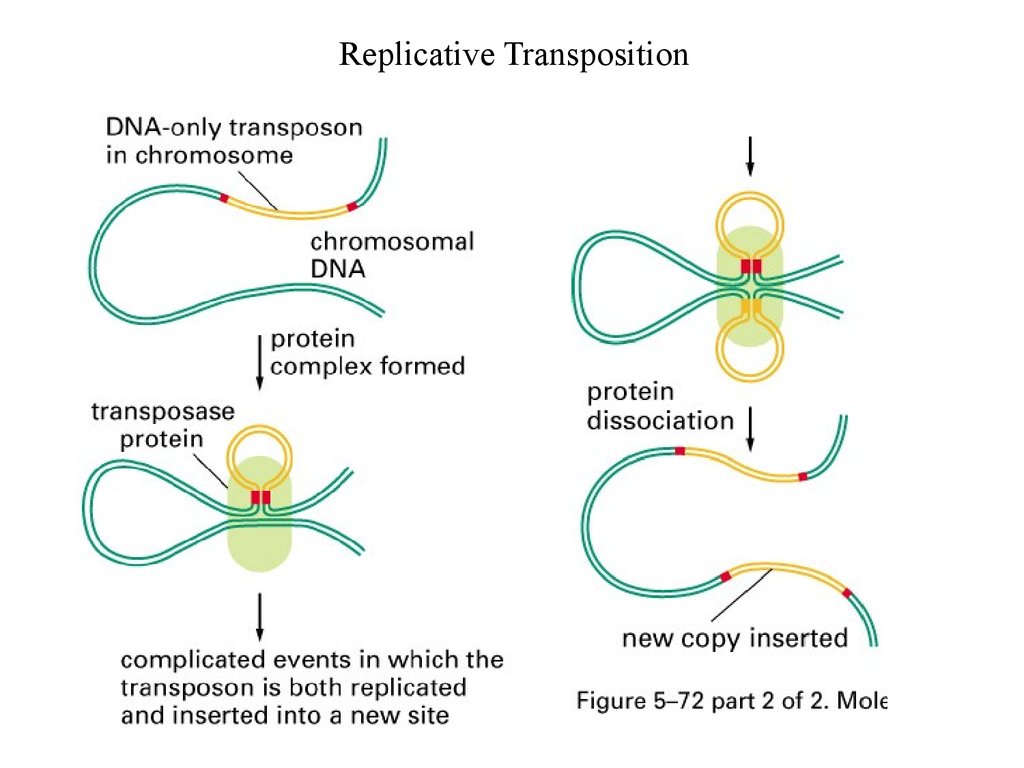
Replicative Transposition9.
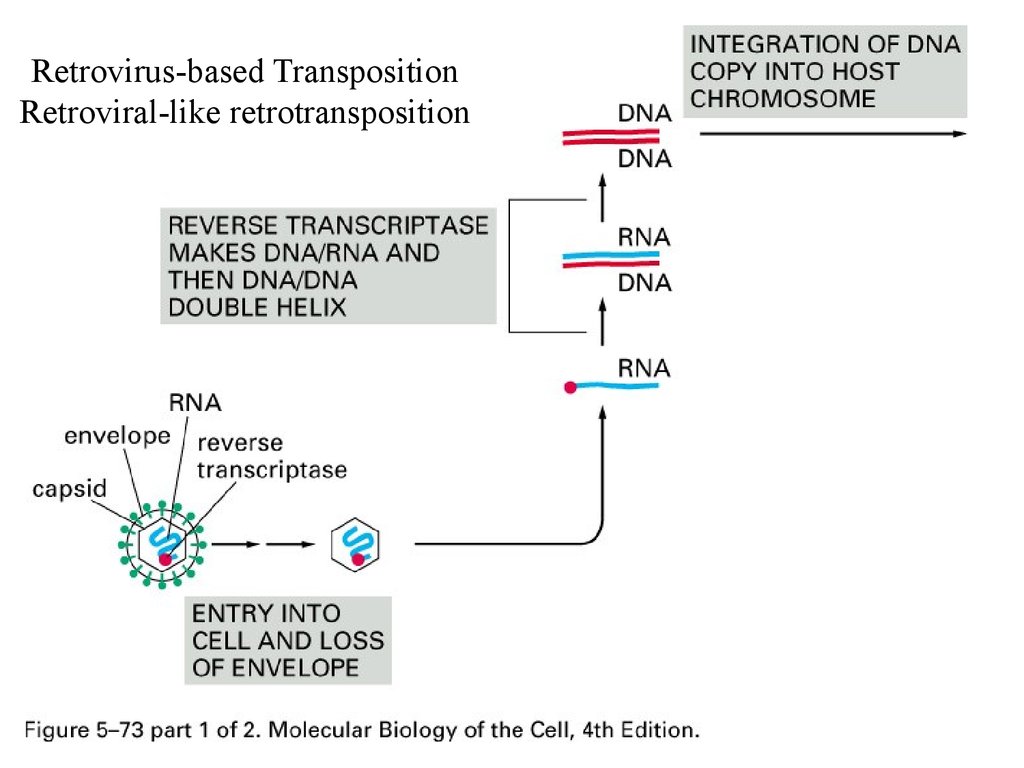
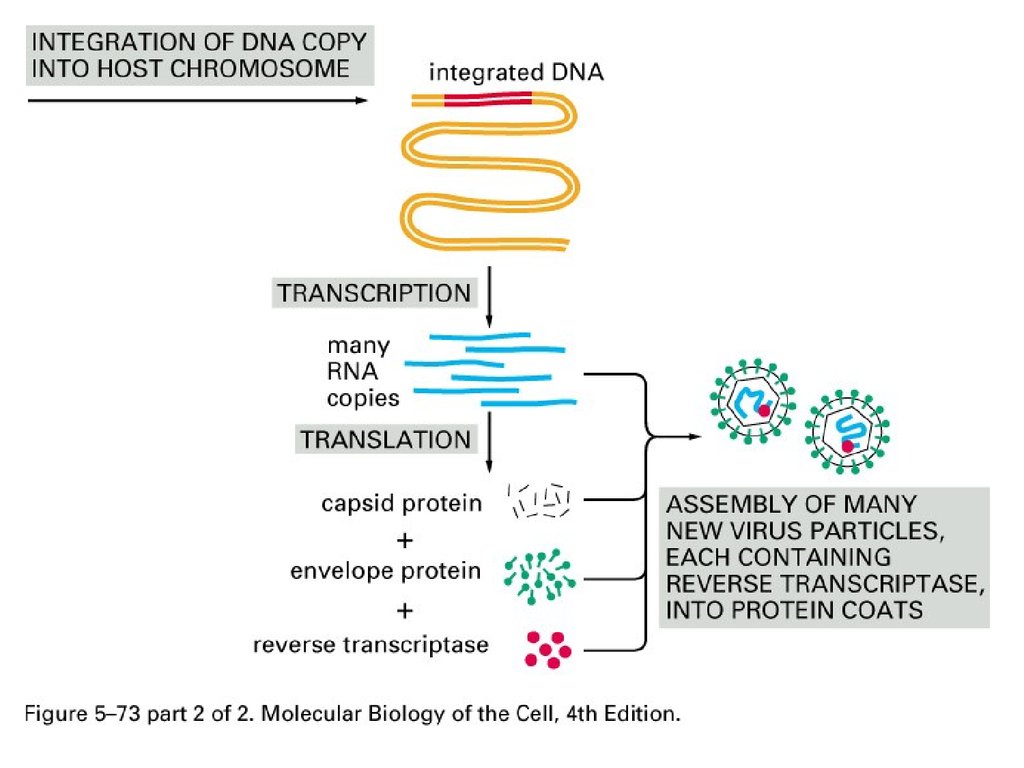
Retrovirus-based TranspositionRetroviral-like retrotransposition
10.
11.
Reverse TranscriptaseFrom RNA to DNA
12.
Non-retroviral retrotranspositionL1 Element
13.
Conservative Site Specific RecombinationIntegration vs. inversion
Notice the arrows of directions
14.
Bacteriophase Lambda15.
Genetic Engineering to control Gene expression16.
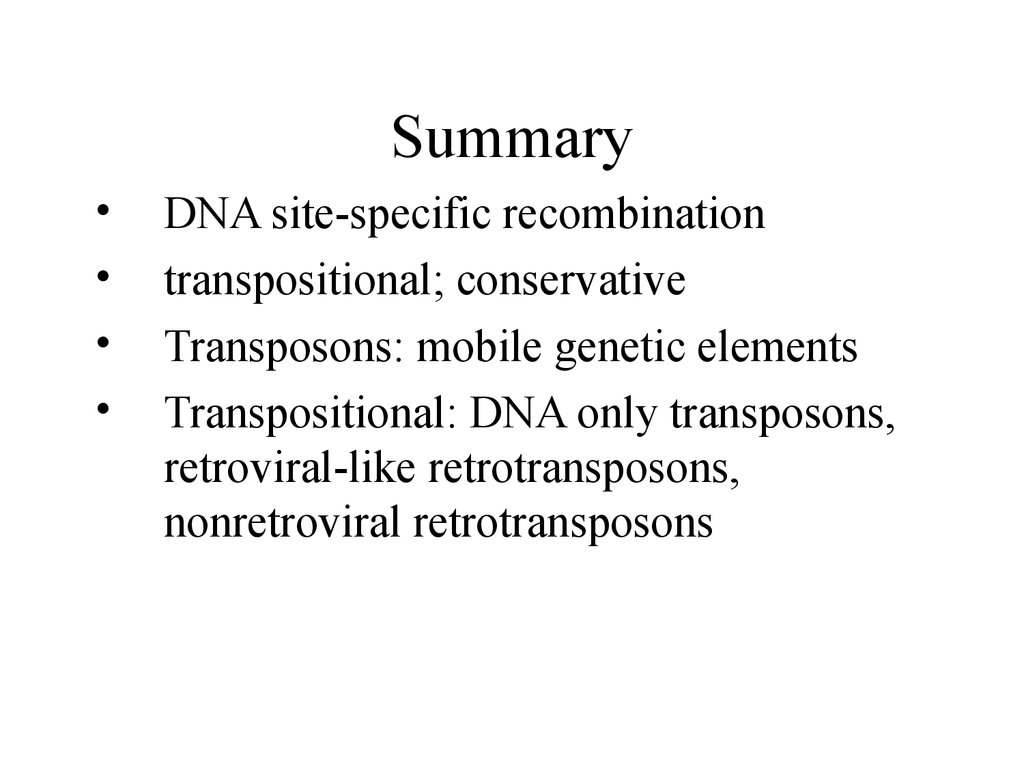
SummaryDNA site-specific recombination
transpositional; conservative
Transposons: mobile genetic elements
Transpositional: DNA only transposons,
retroviral-like retrotransposons,
nonretroviral retrotransposons
17.
• How Cells Read the Genome:From DNA to Protein
1. Transcription
2. RNA Modification and Splicing
3. RNA transportation
4. Translation
5. Protein Modification and
Folding
18.
DNA->RNA-> Proteins19.
Genes expressed with different efficiency20.
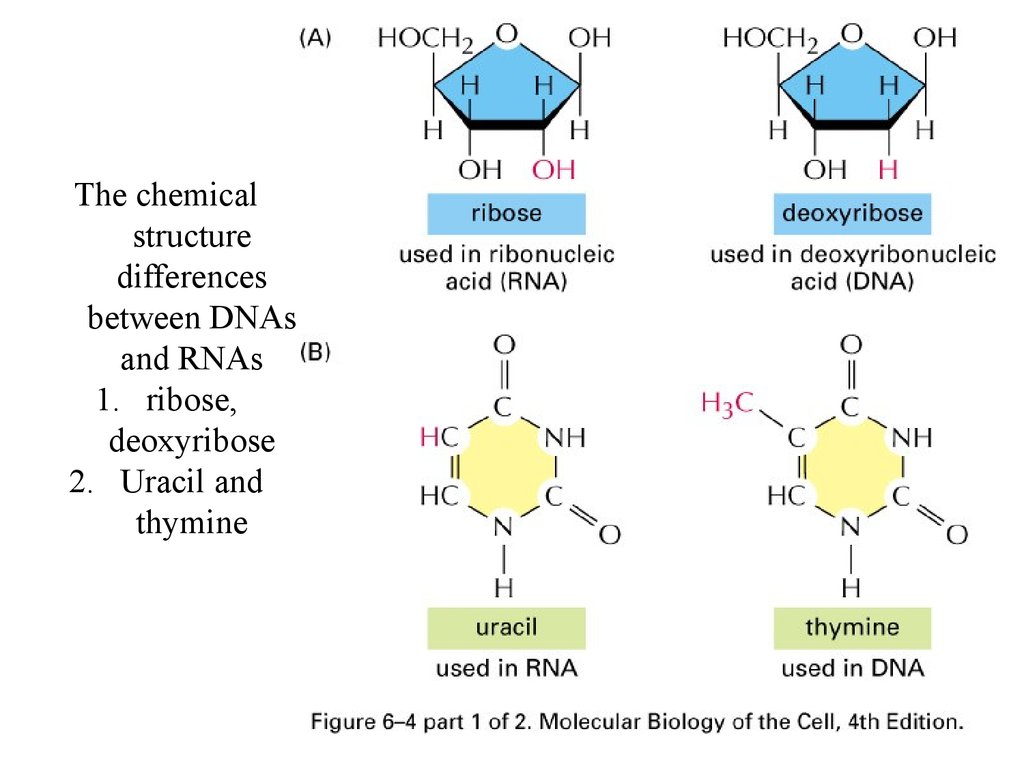
The chemicalstructure
differences
between DNAs
and RNAs
1. ribose,
deoxyribose
2. Uracil and
thymine
21.
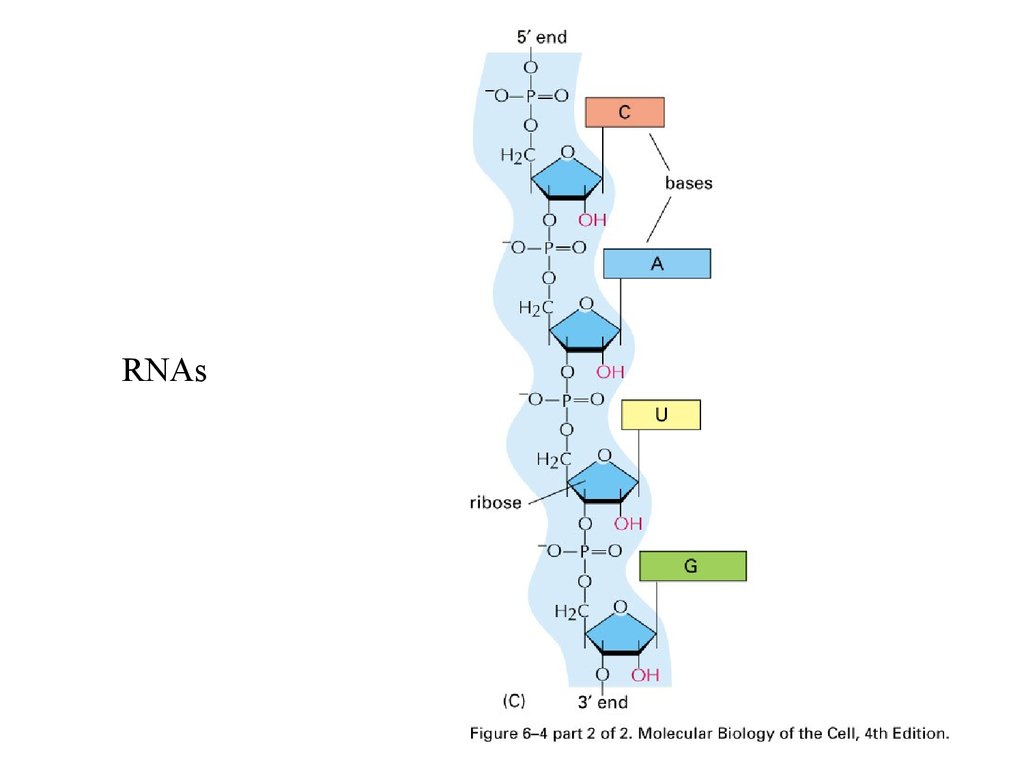
RNAs22.
RNA base pairsA-U; G-C
23.
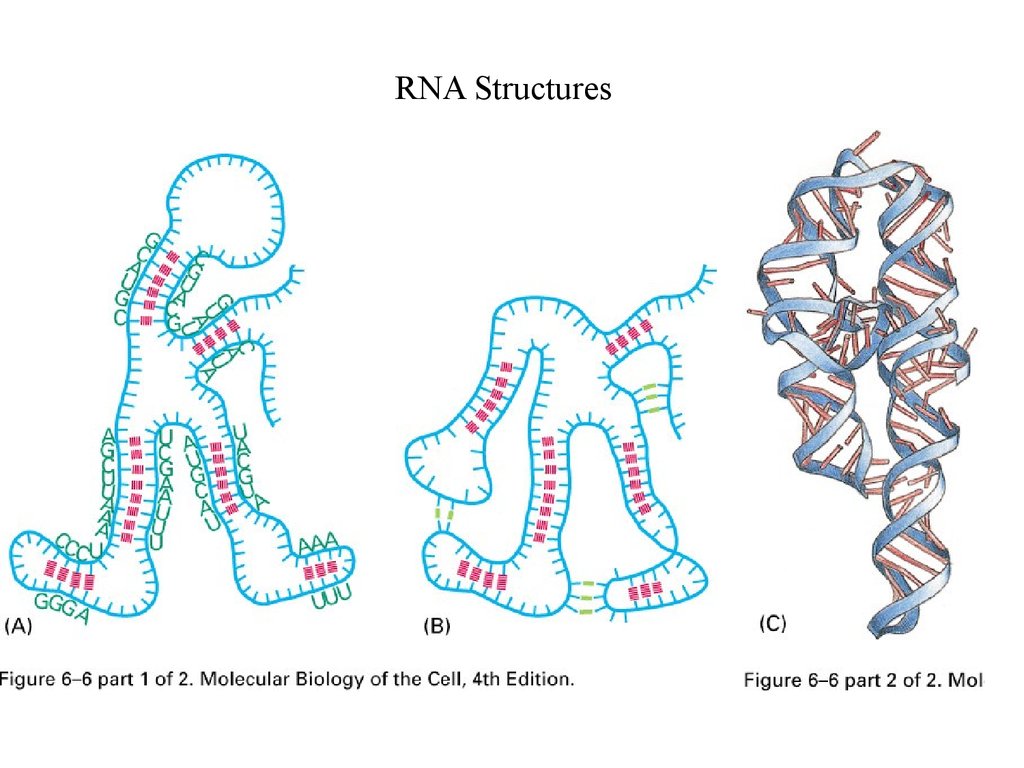
RNA Structures24.
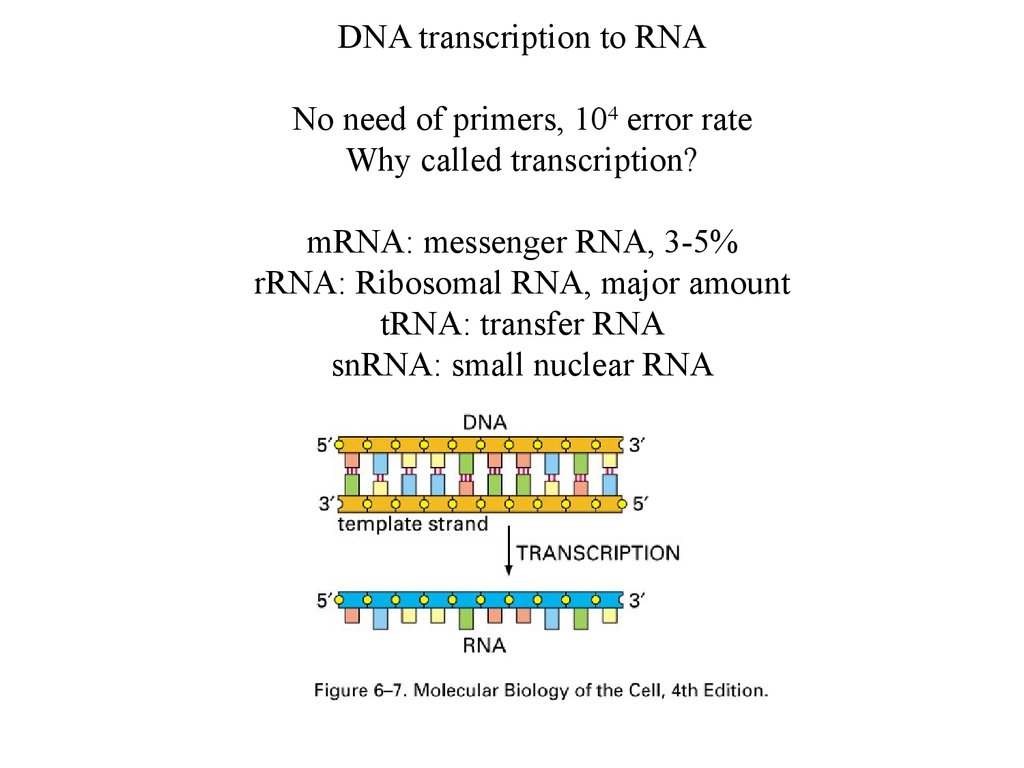
DNA transcription to RNANo need of primers, 104 error rate
Why called transcription?
mRNA: messenger RNA, 3-5%
rRNA: Ribosomal RNA, major amount
tRNA: transfer RNA
snRNA: small nuclear RNA
25.
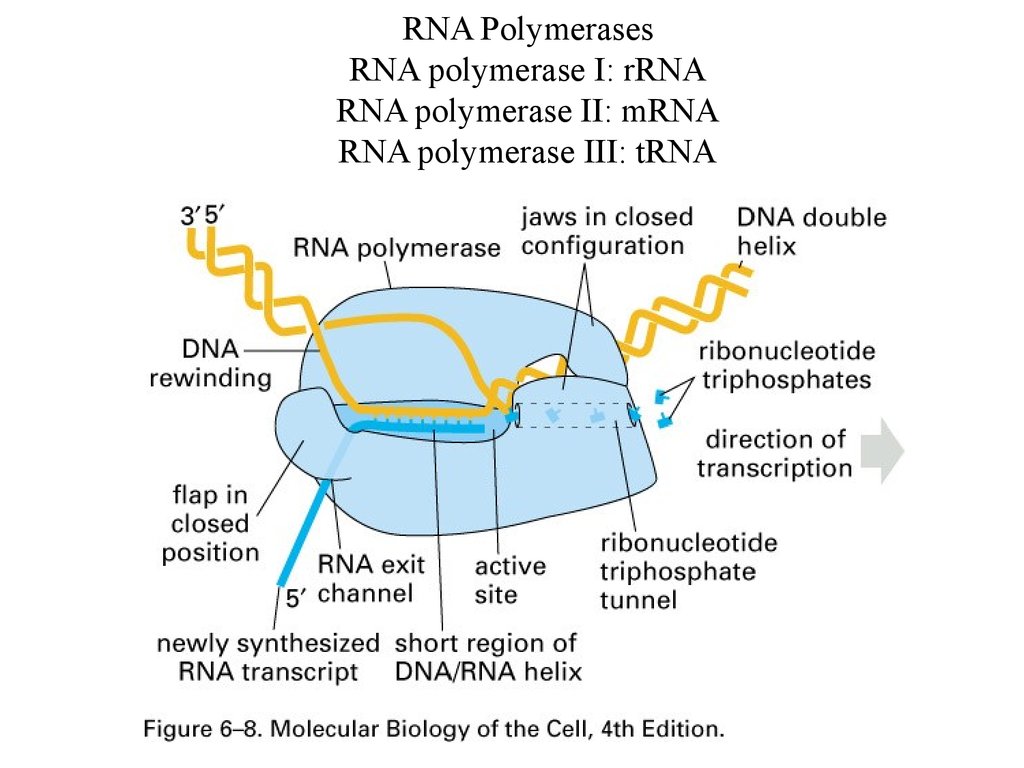
RNA PolymerasesRNA polymerase I: rRNA
RNA polymerase II: mRNA
RNA polymerase III: tRNA
26.
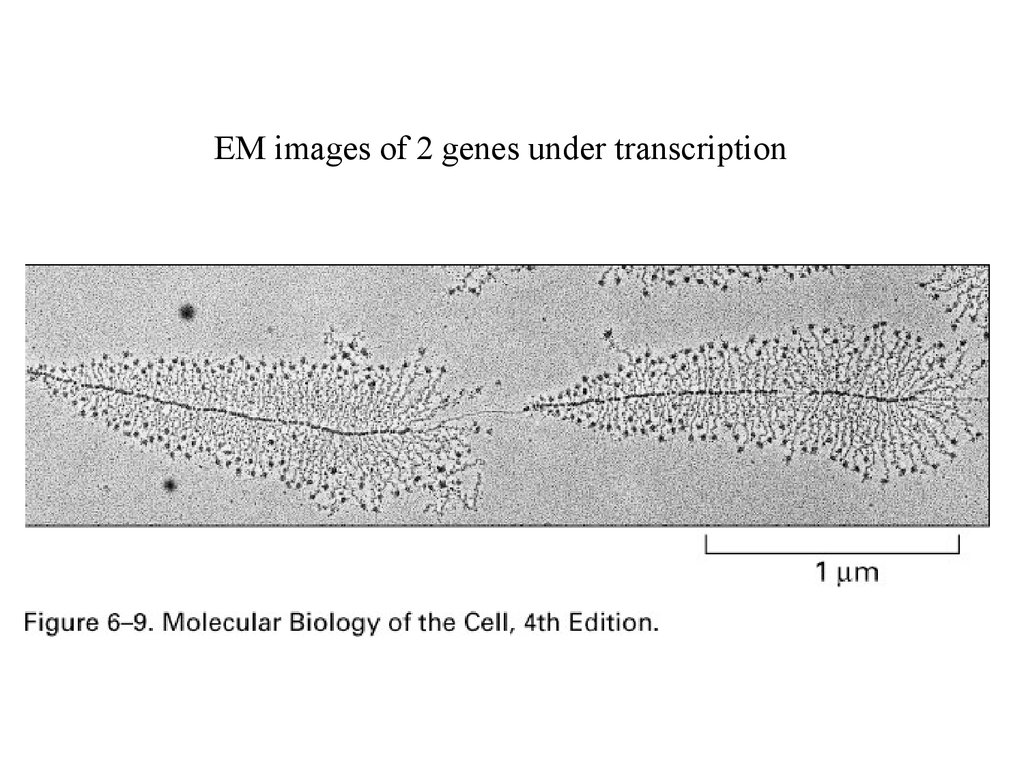
EM images of 2 genes under transcription27.
TranscriptionCycle
Promoter
Terminator
sigma factor
28.
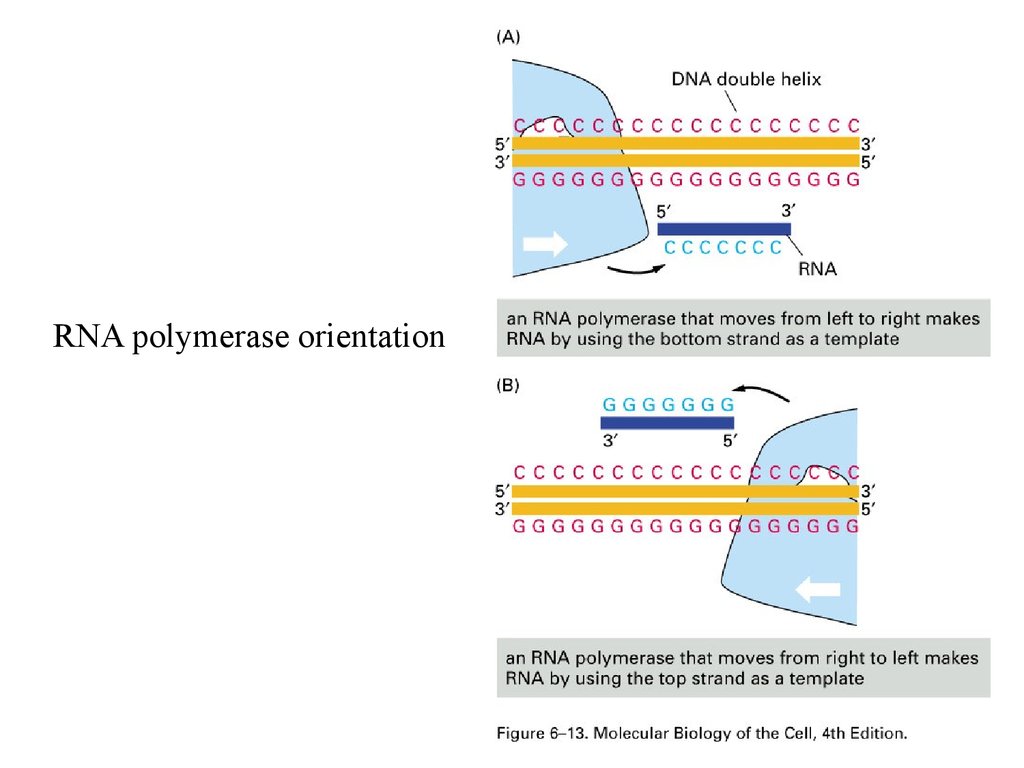
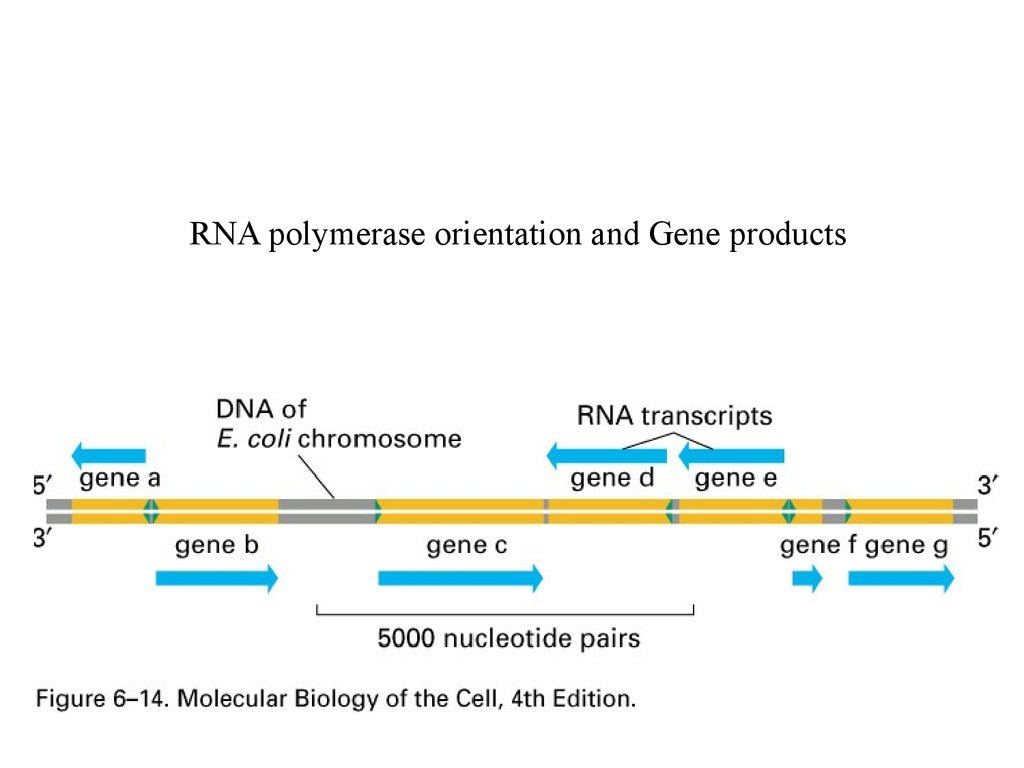
RNA polymerase orientation29.
RNA polymerase orientation and Gene products30.
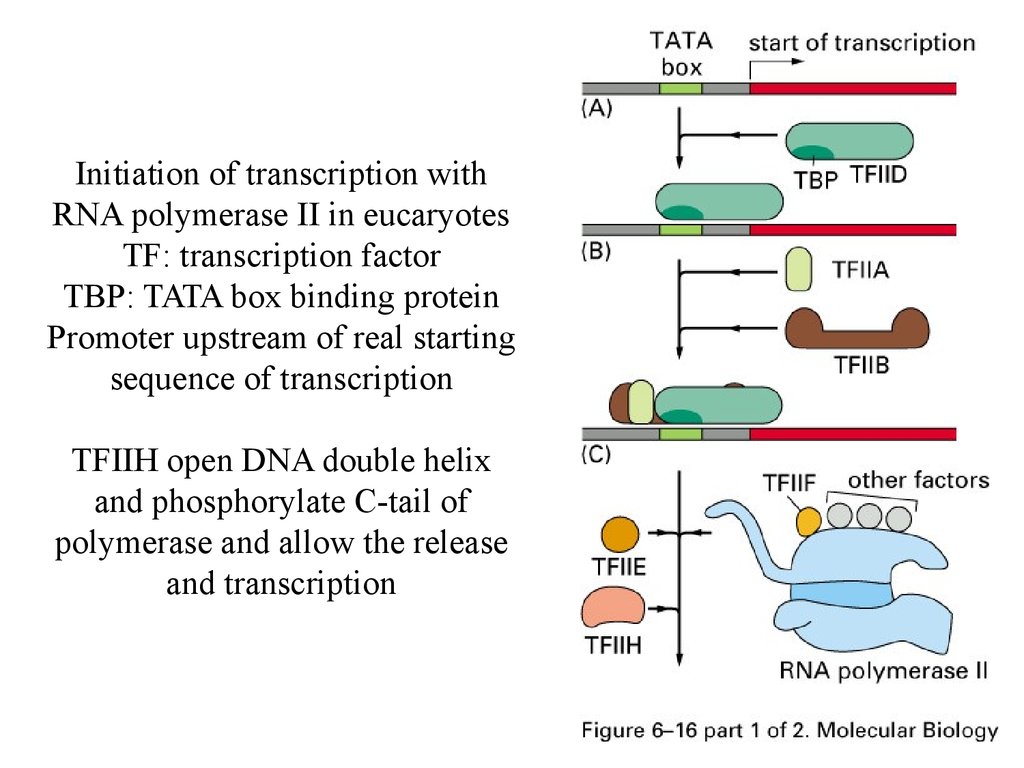
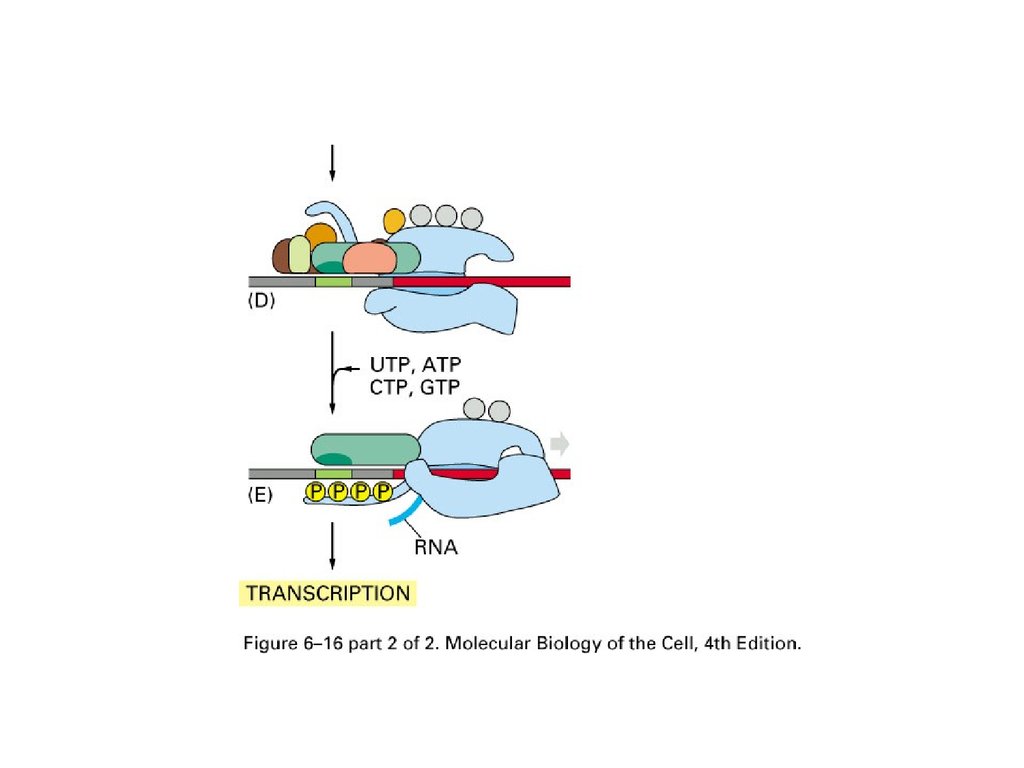
Initiation of transcription withRNA polymerase II in eucaryotes
TF: transcription factor
TBP: TATA box binding protein
Promoter upstream of real starting
sequence of transcription
TFIIH open DNA double helix
and phosphorylate C-tail of
polymerase and allow the release
and transcription
31.
32.
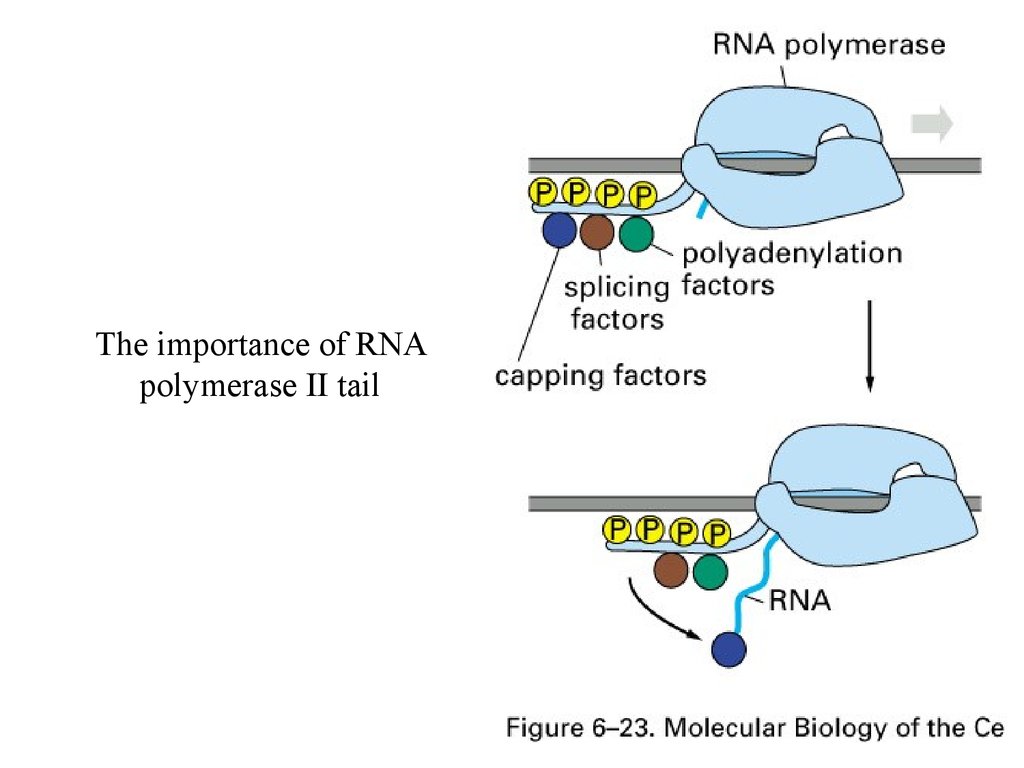
The importance of RNApolymerase II tail
33.
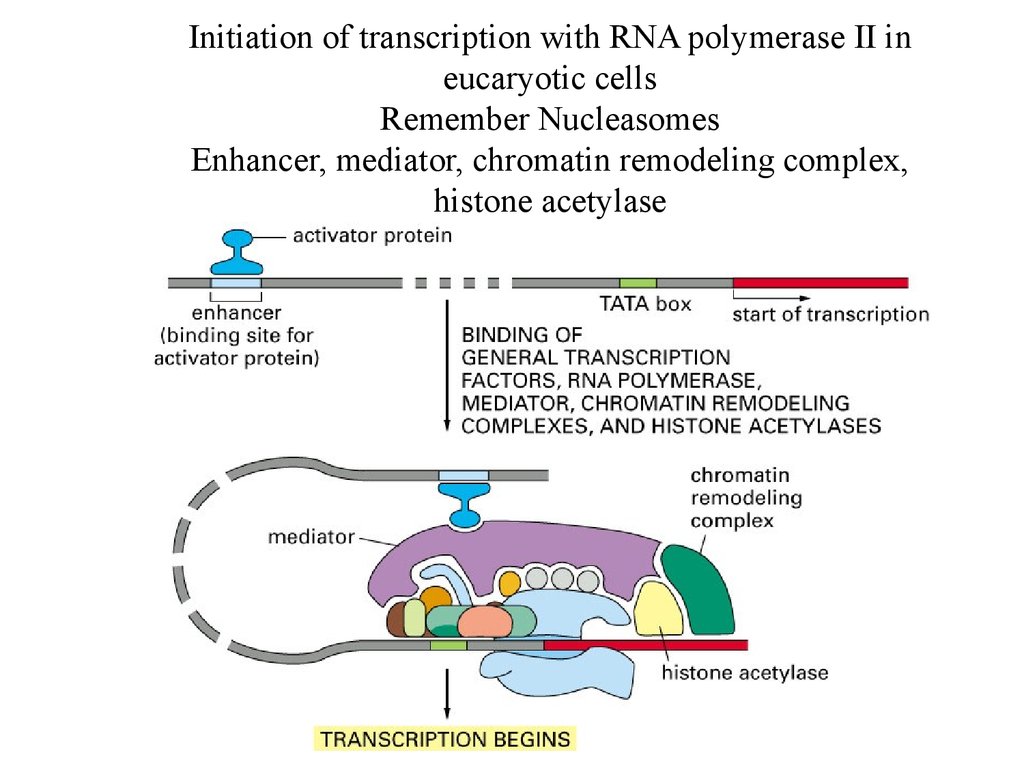
Initiation of transcription with RNA polymerase II ineucaryotic cells
Remember Nucleasomes
Enhancer, mediator, chromatin remodeling complex,
histone acetylase
34.
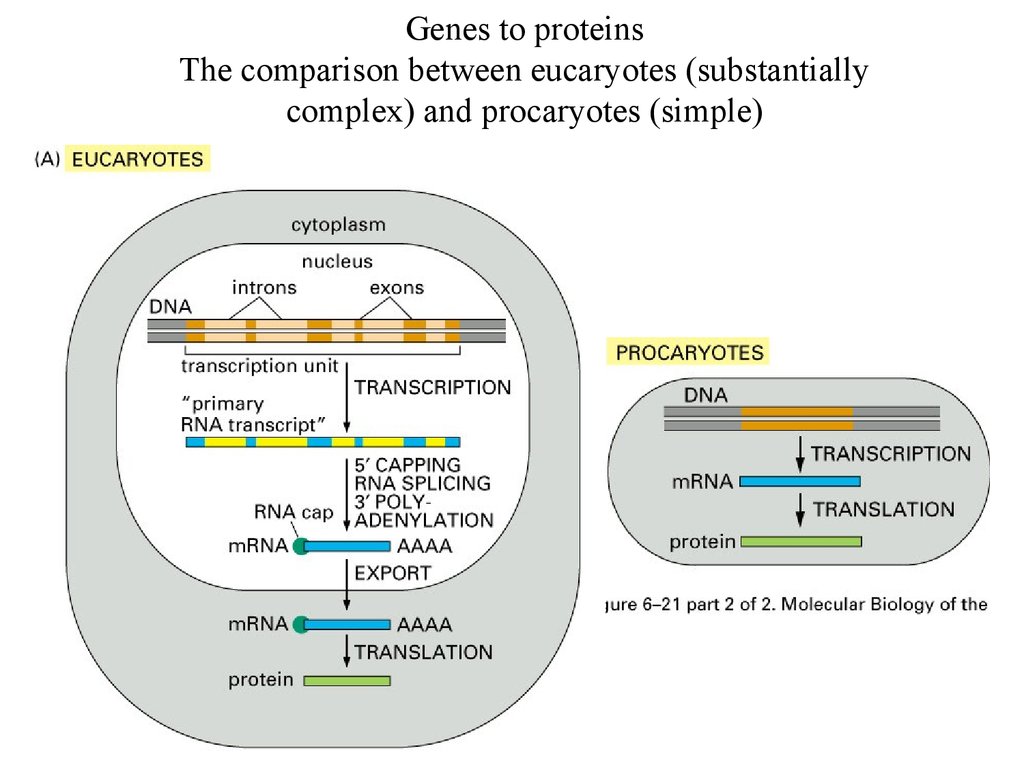
Genes to proteinsThe comparison between eucaryotes (substantially
complex) and procaryotes (simple)
35.
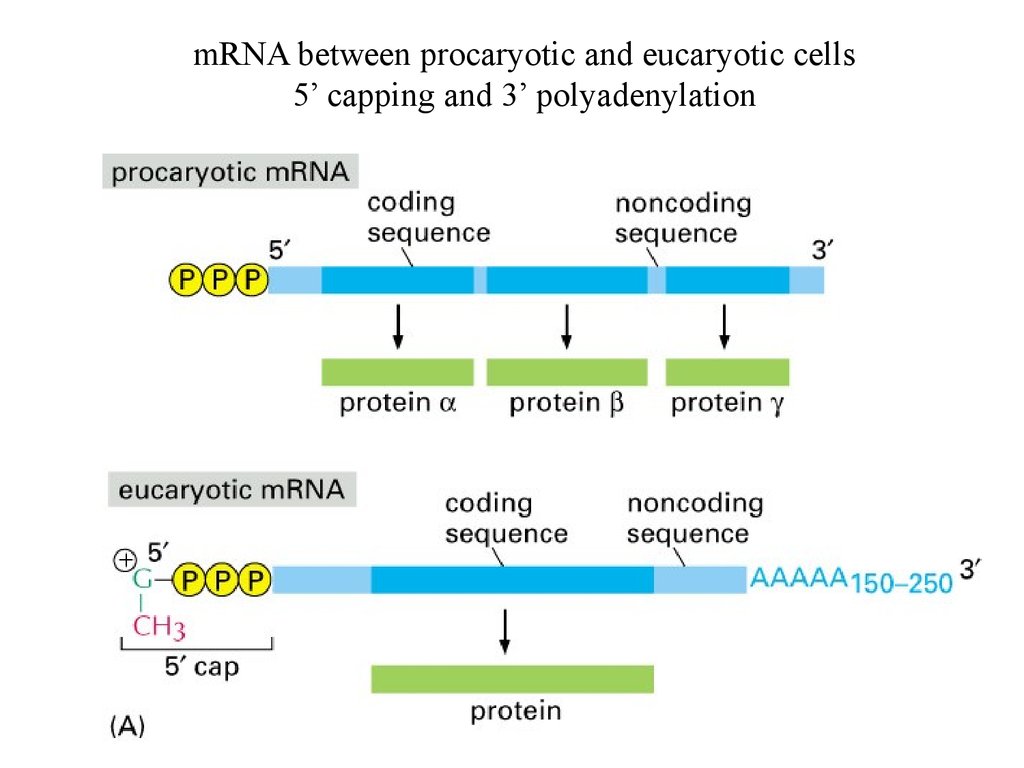
mRNA between procaryotic and eucaryotic cells5’ capping and 3’ polyadenylation
36.
5’ capping37.
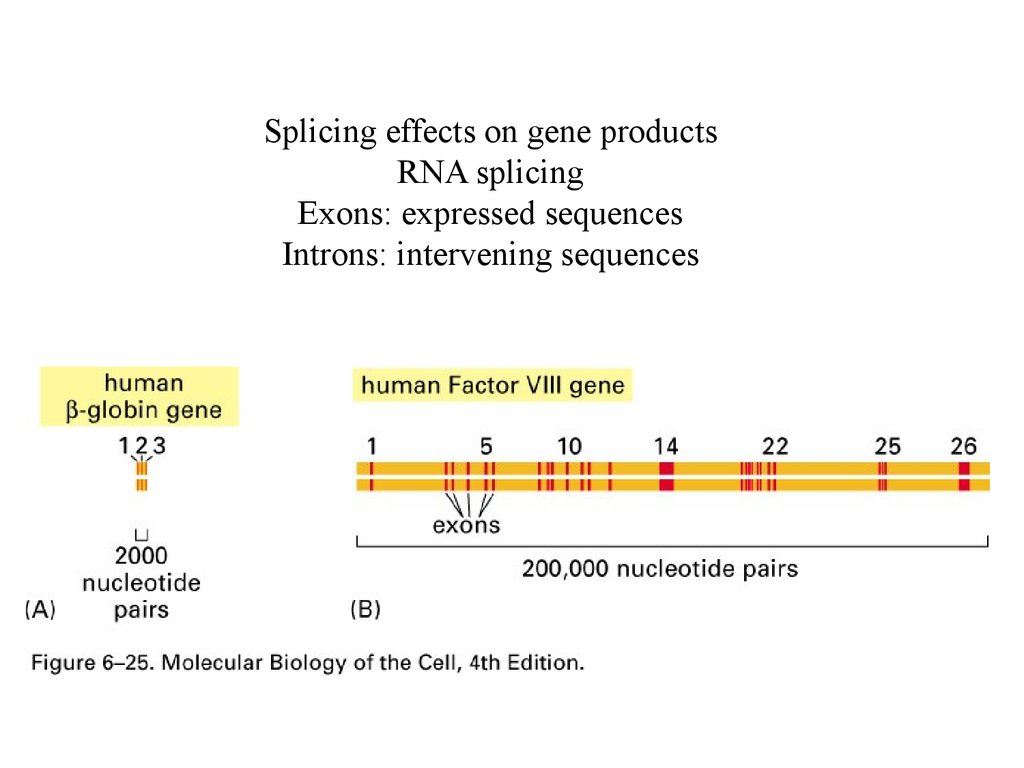
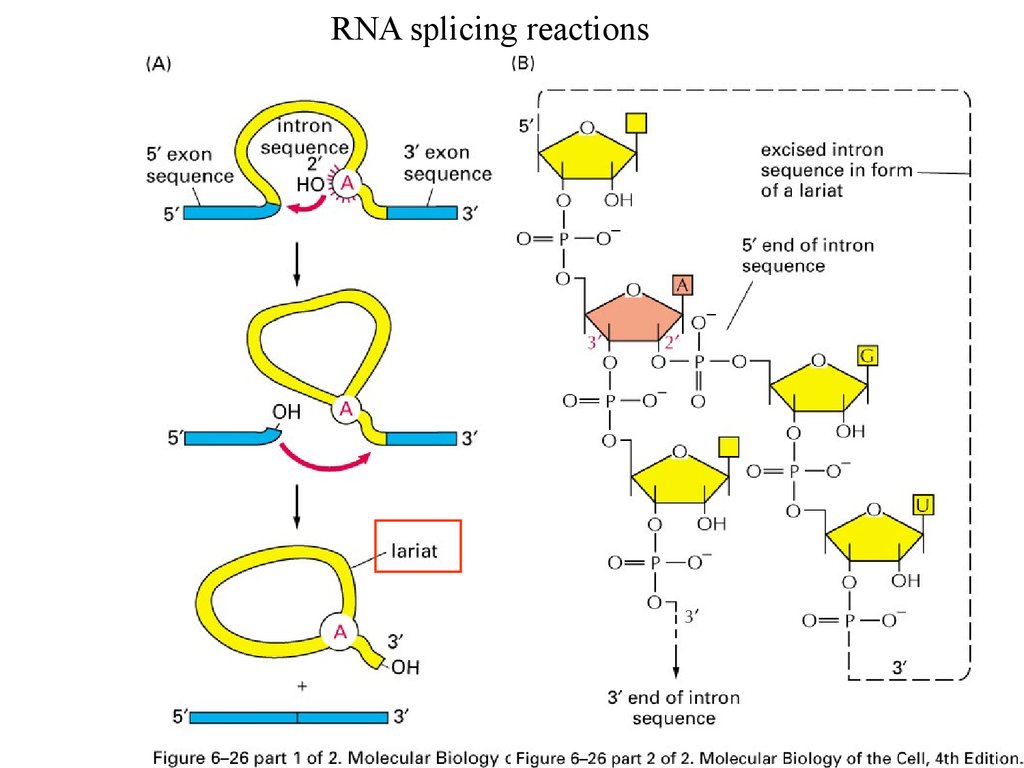
Splicing effects on gene productsRNA splicing
Exons: expressed sequences
Introns: intervening sequences
38.
RNA splicing reactions39.
3 Important sequences for Splicing to occurR: A or G; Y: C or U
40.
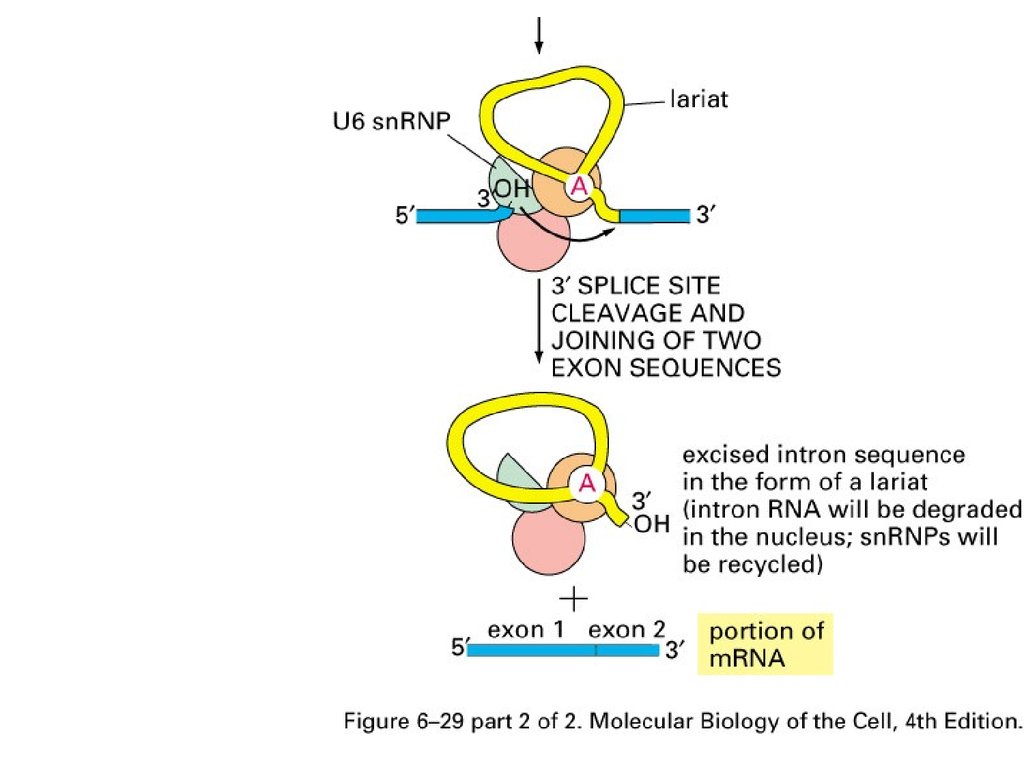
RNA Splicing mechanismBBP: branch-point binding
protein
U2AF: a helper protein
snRNA: small nuclear RNA
snRNP: small nuclear
ribonucleoprotein
Components for splicesome
41.
42.
Further mechanism to mark Exon and Intron differenceCBC: capping binding complex
hnRNP: heterogeneous nuclear ribonucleoprotein, binding
to introns
SR: rich in serine and arginines, binding to exons
43.
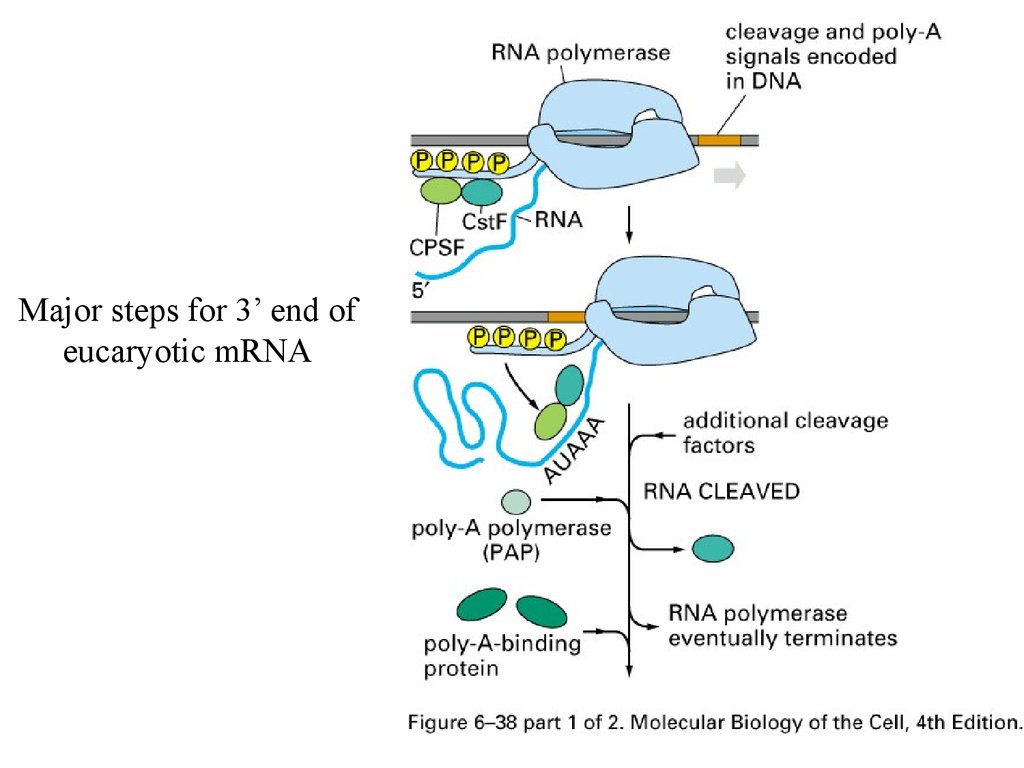
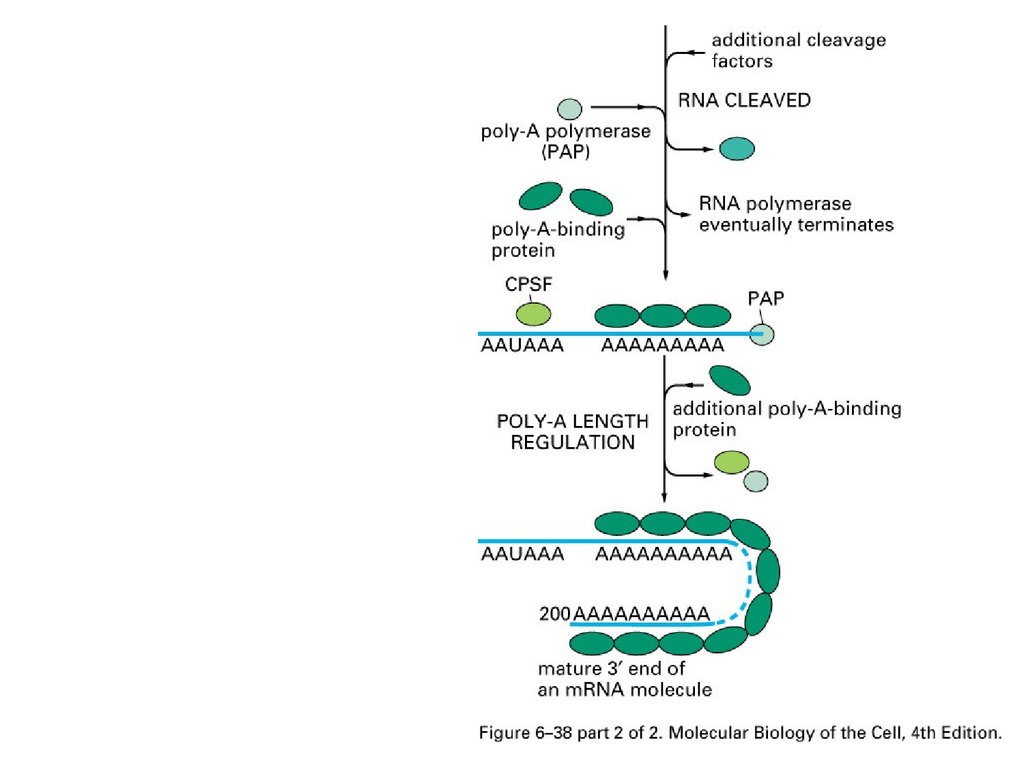
Consensus sequence for 3’ processAAUAAA: CstF (cleavage stimulation factor F)
GU-rich sequence: CPSF (cleavage and polyadenylation specificity factor)
44.
Major steps for 3’ end ofeucaryotic mRNA
45.
46.
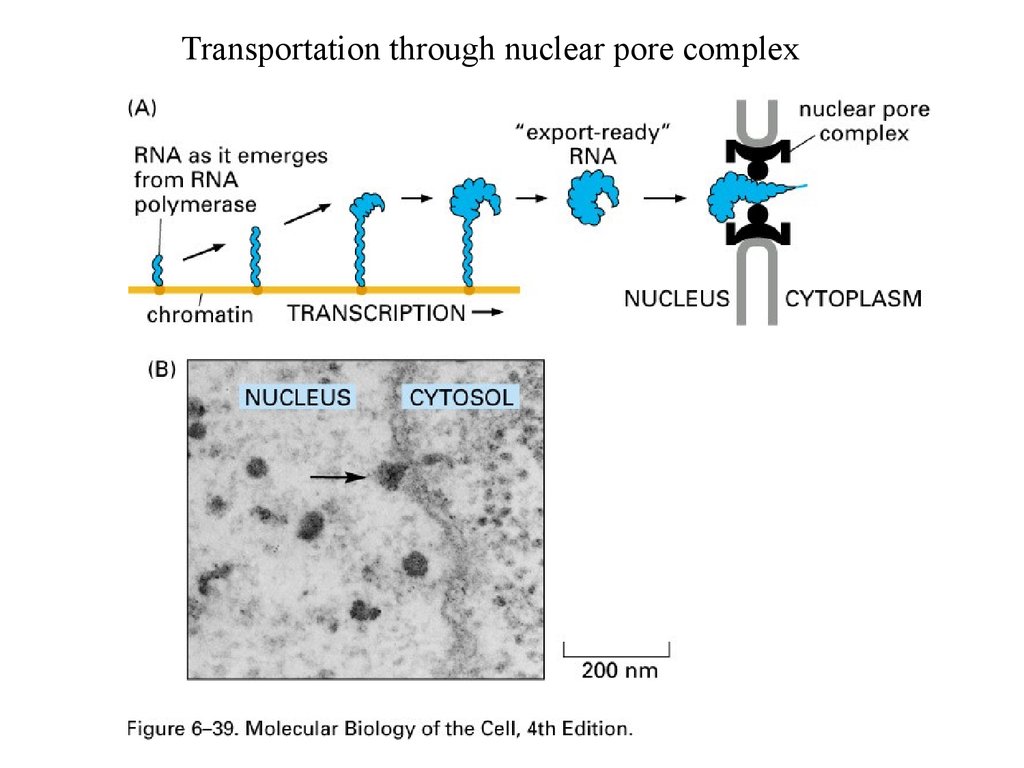
Transportation through nuclear pore complex47.
Exporting mechanismhnRNP binds to intron and help the recognition to destroy RNA introns
48.
49.
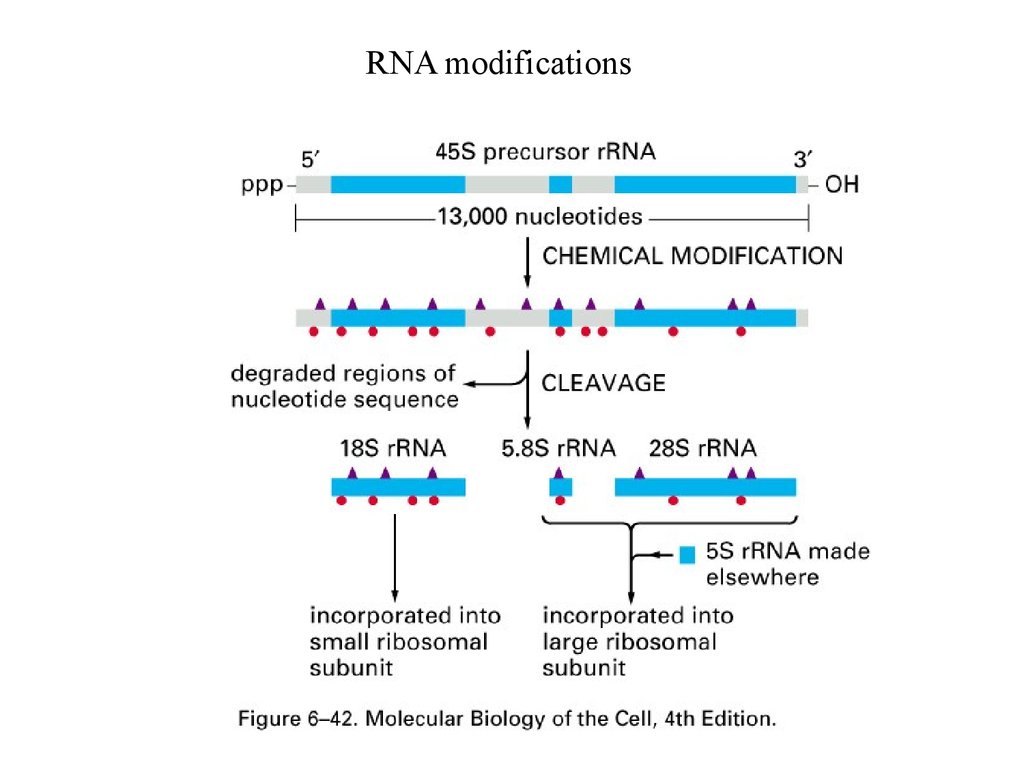
RNA modifications50.
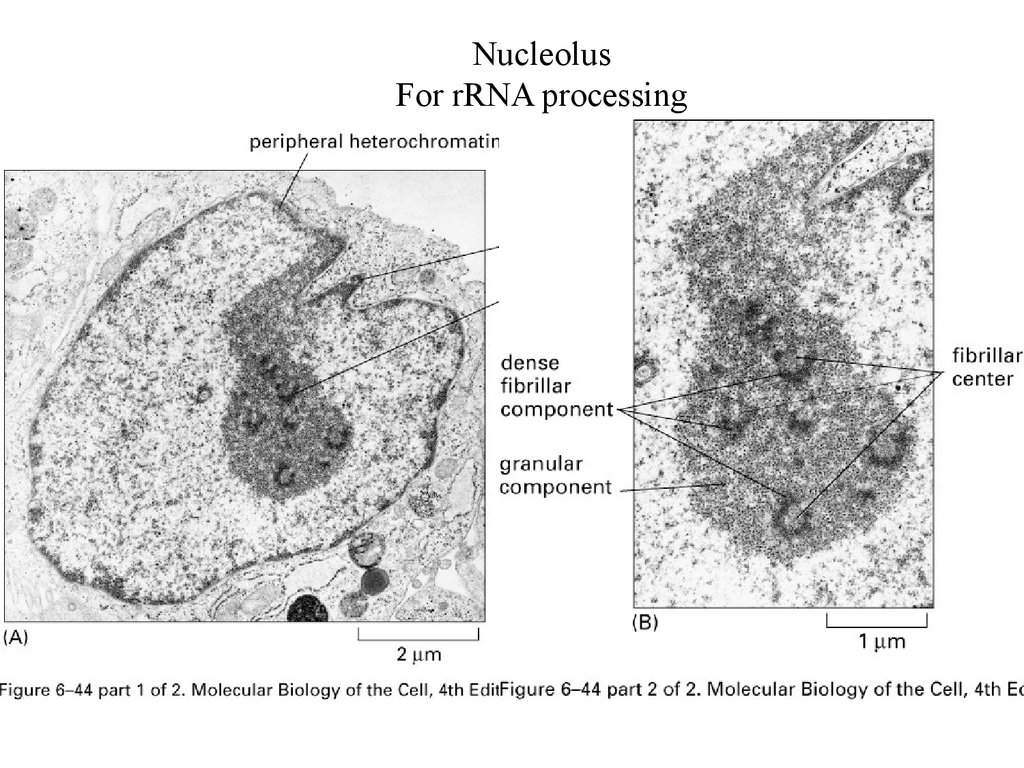
NucleolusFor rRNA processing
51.
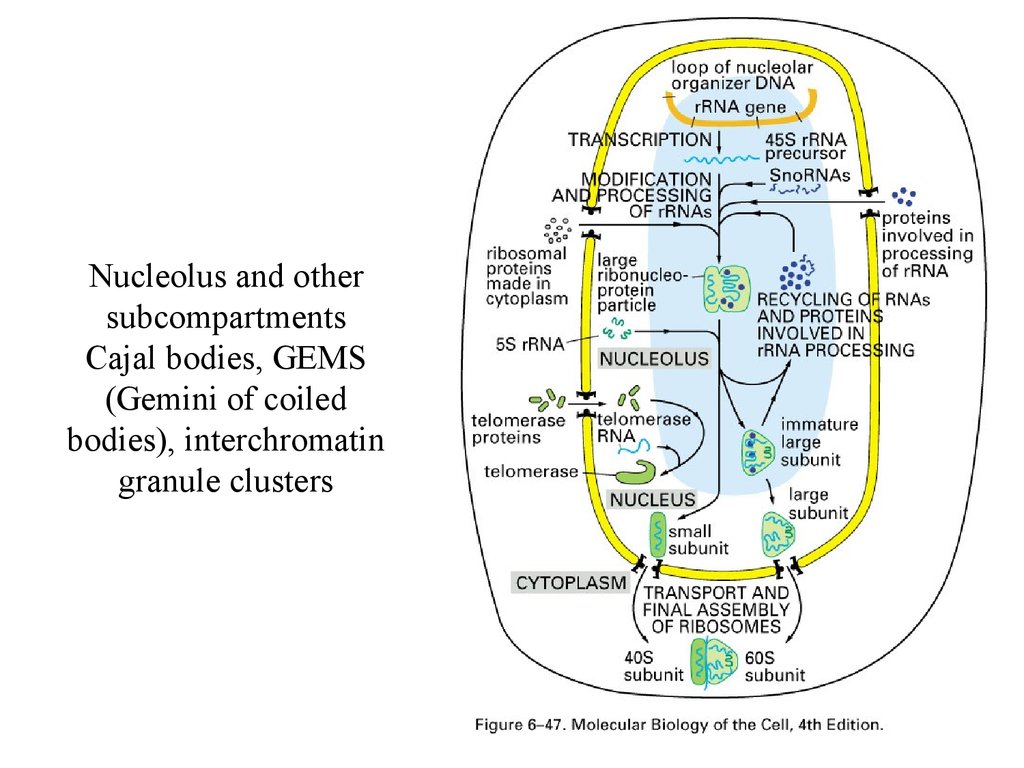
Nucleolus and othersubcompartments
Cajal bodies, GEMS
(Gemini of coiled
bodies), interchromatin
granule clusters
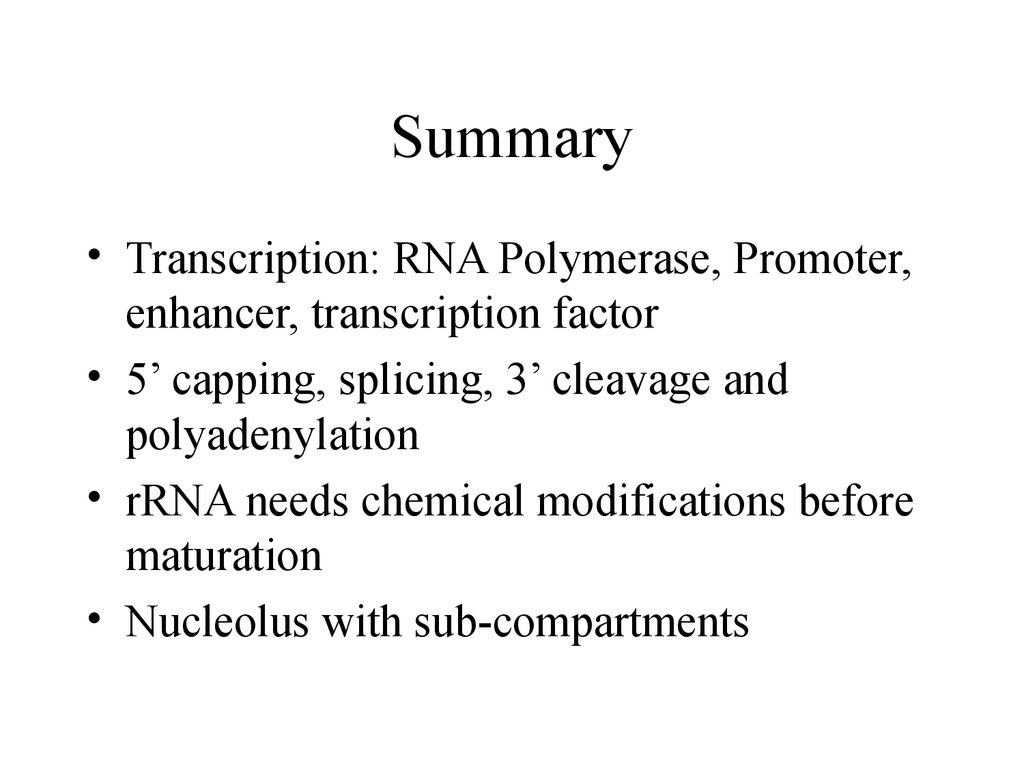
52. Summary
• Transcription: RNA Polymerase, Promoter,enhancer, transcription factor
• 5’ capping, splicing, 3’ cleavage and
polyadenylation
• rRNA needs chemical modifications before
maturation
• Nucleolus with sub-compartments
53.
• From RNA to Protein1. Protein synthesis
2. Protein Folding and regulation
54.
The Genetic Code55.
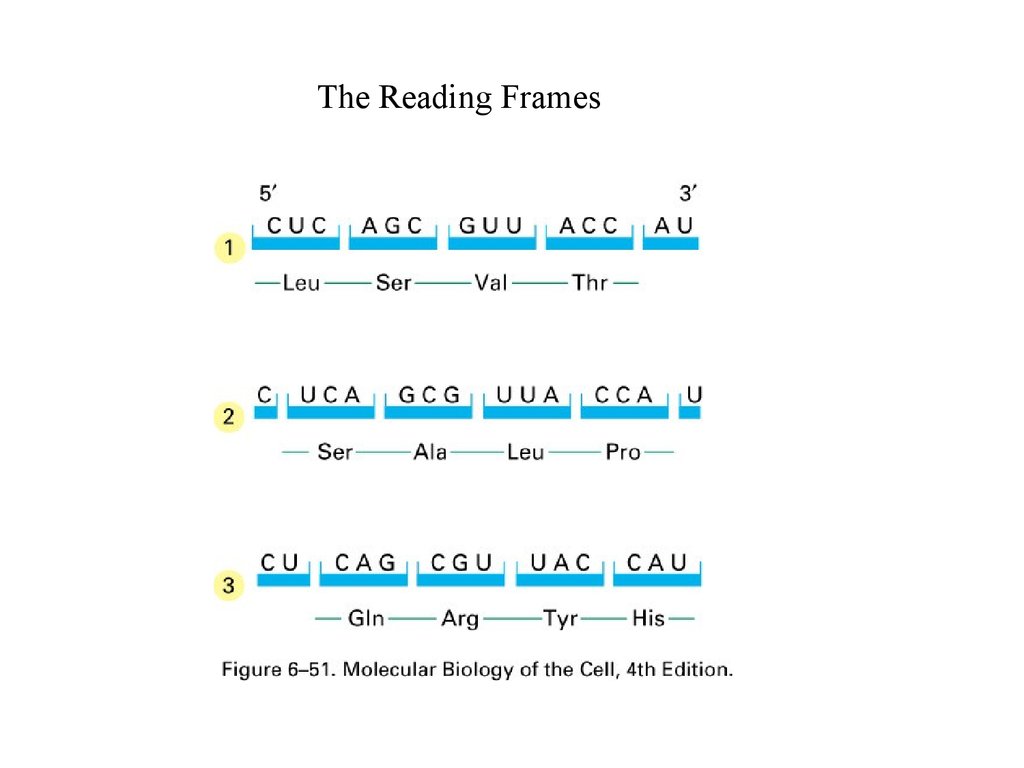
The Reading Frames56.
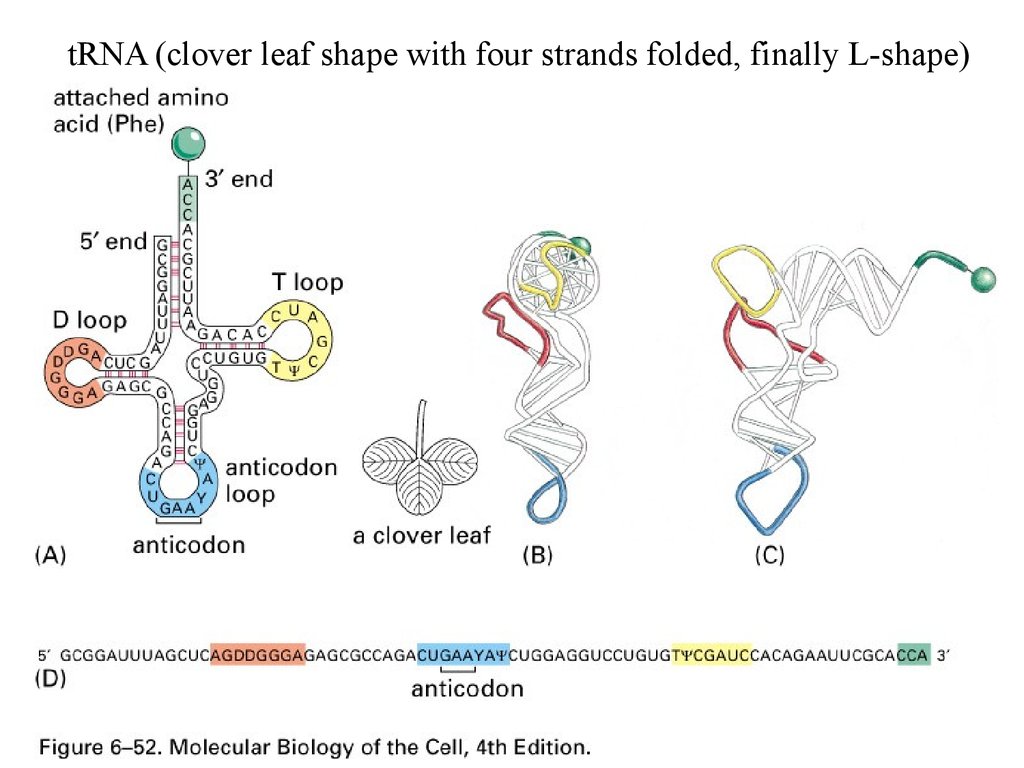
tRNA (clover leaf shape with four strands folded, finally L-shape)57.
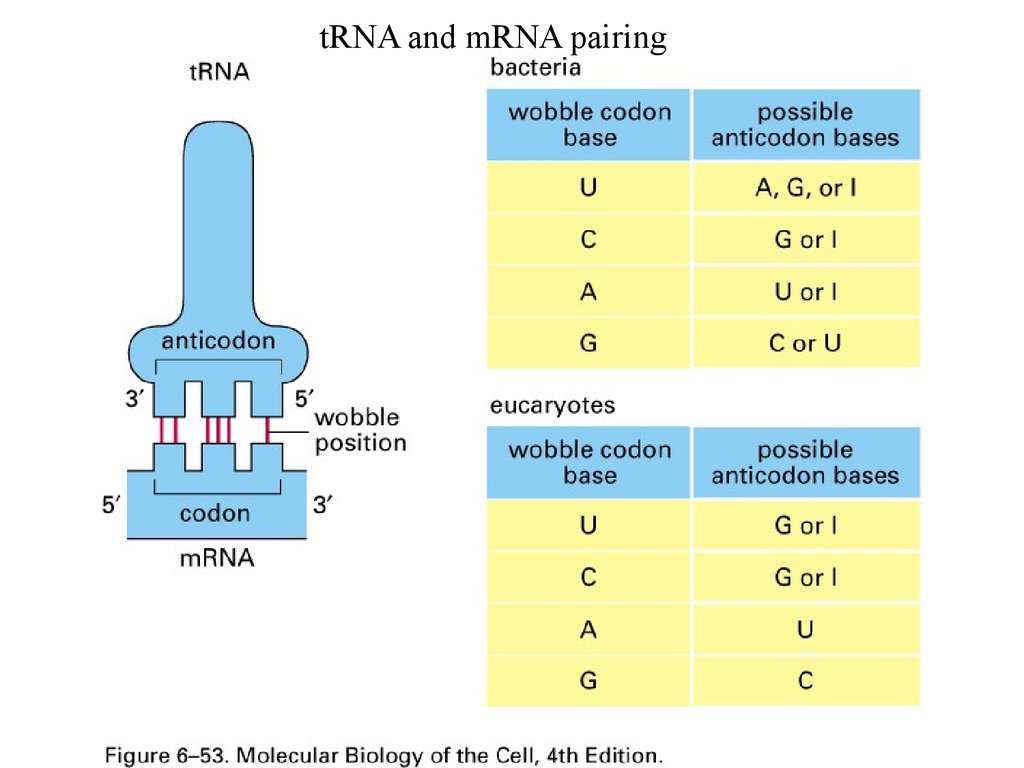
tRNA and mRNA pairing58.
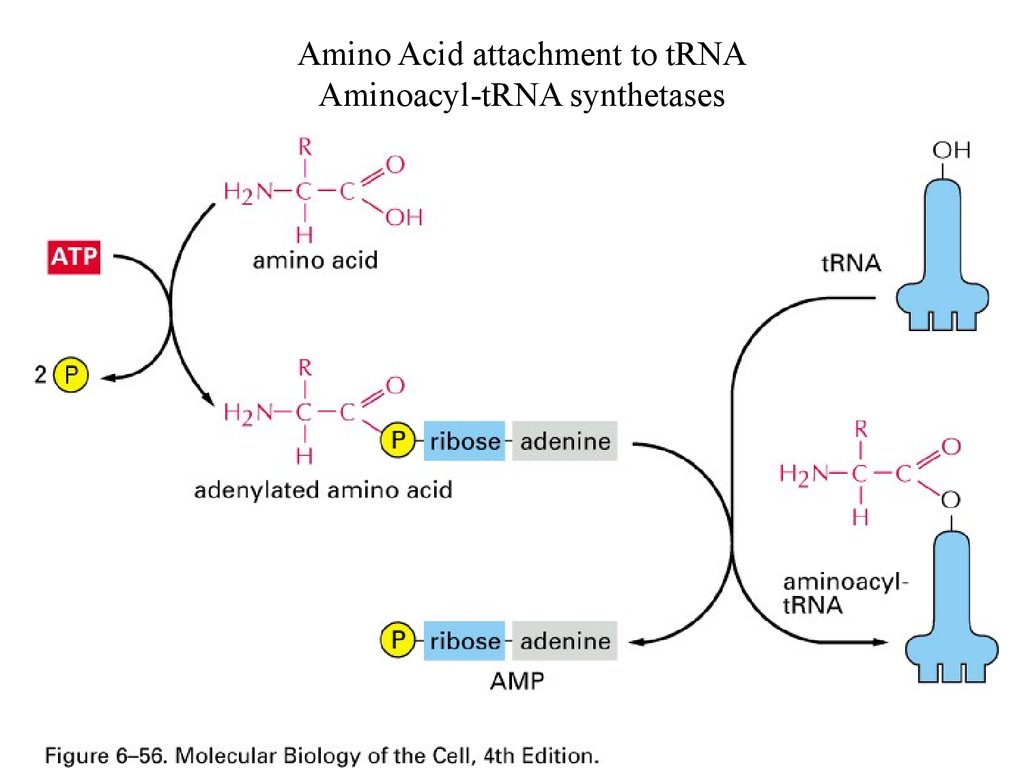
Amino Acid attachment to tRNAAminoacyl-tRNA synthetases
59.
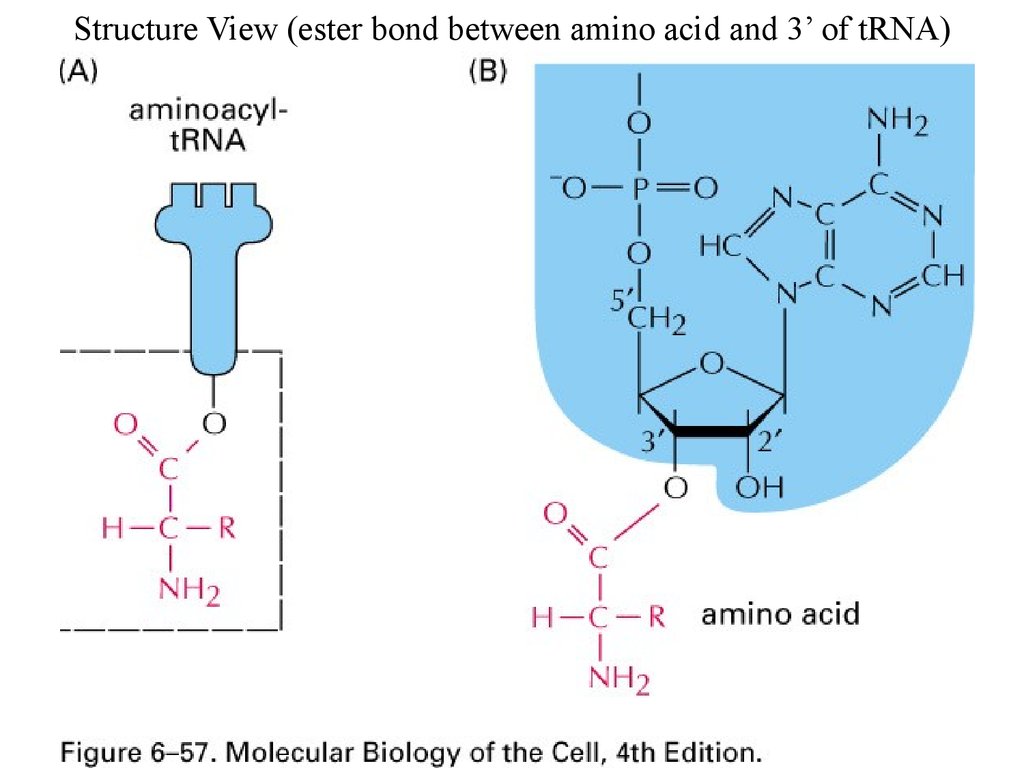
Structure View (ester bond between amino acid and 3’ of tRNA)60.
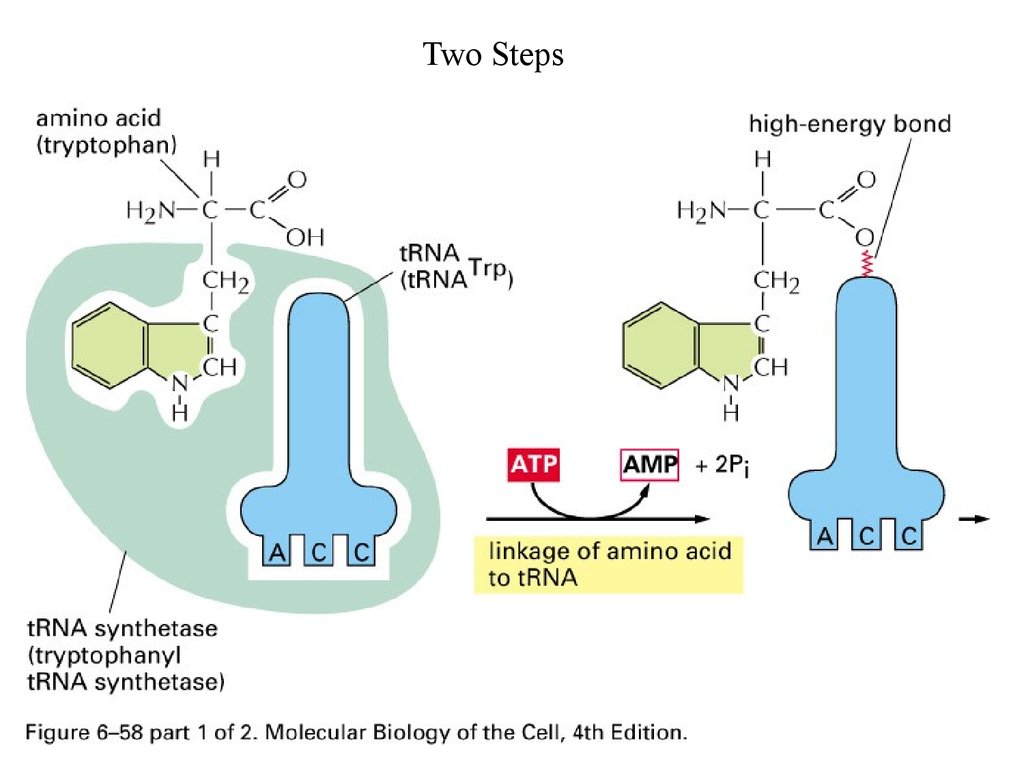
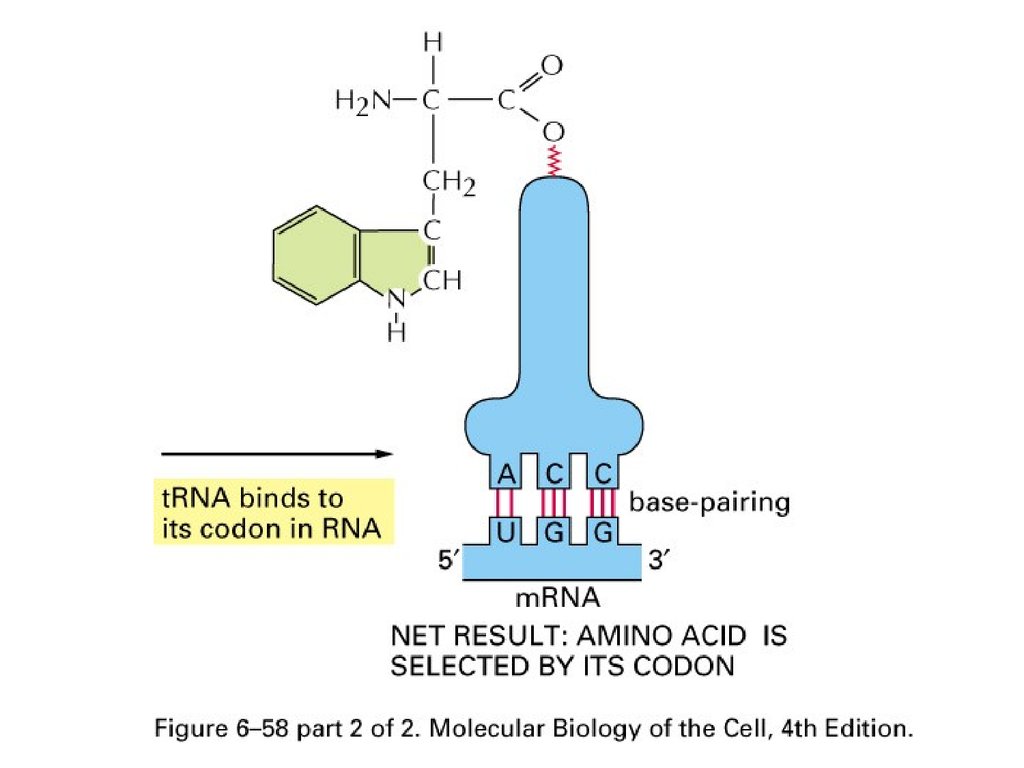
Two Steps61.
62.
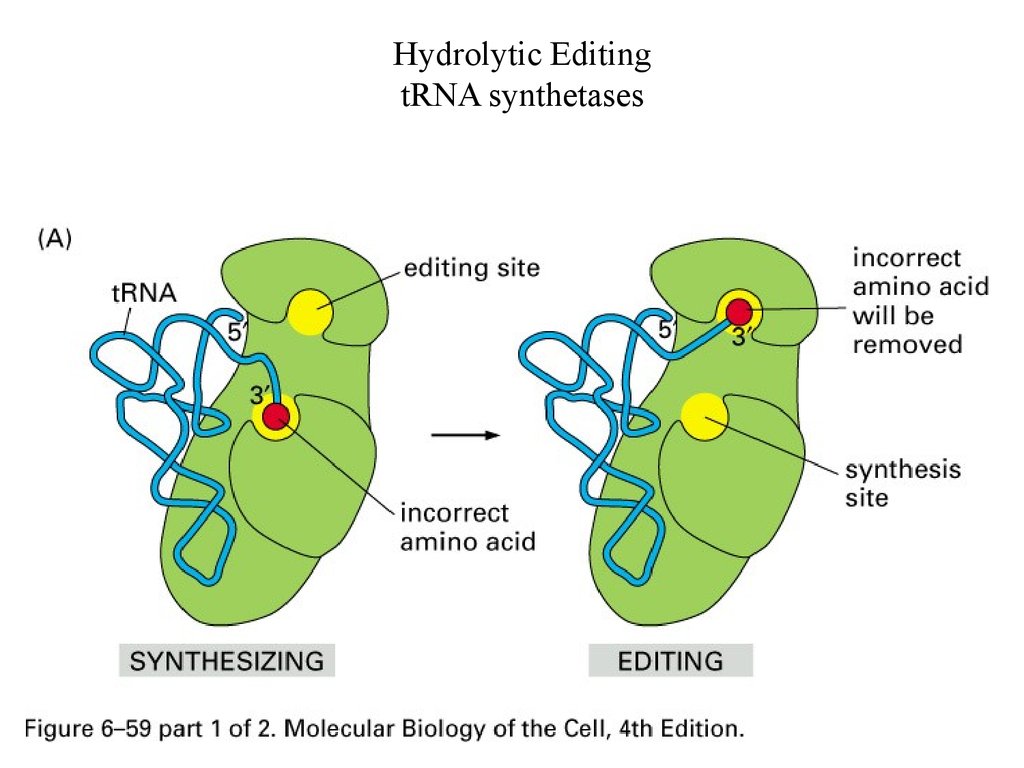
Hydrolytic EditingtRNA synthetases
63.
Hydrolytic EditingDNA polymerase
64.
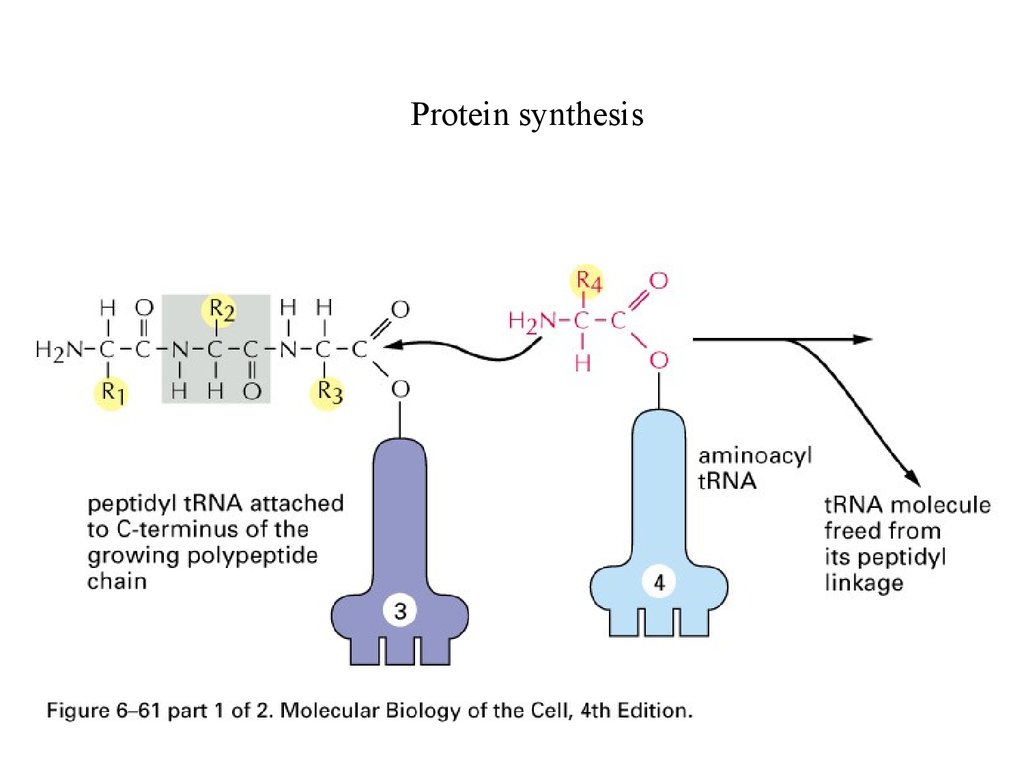
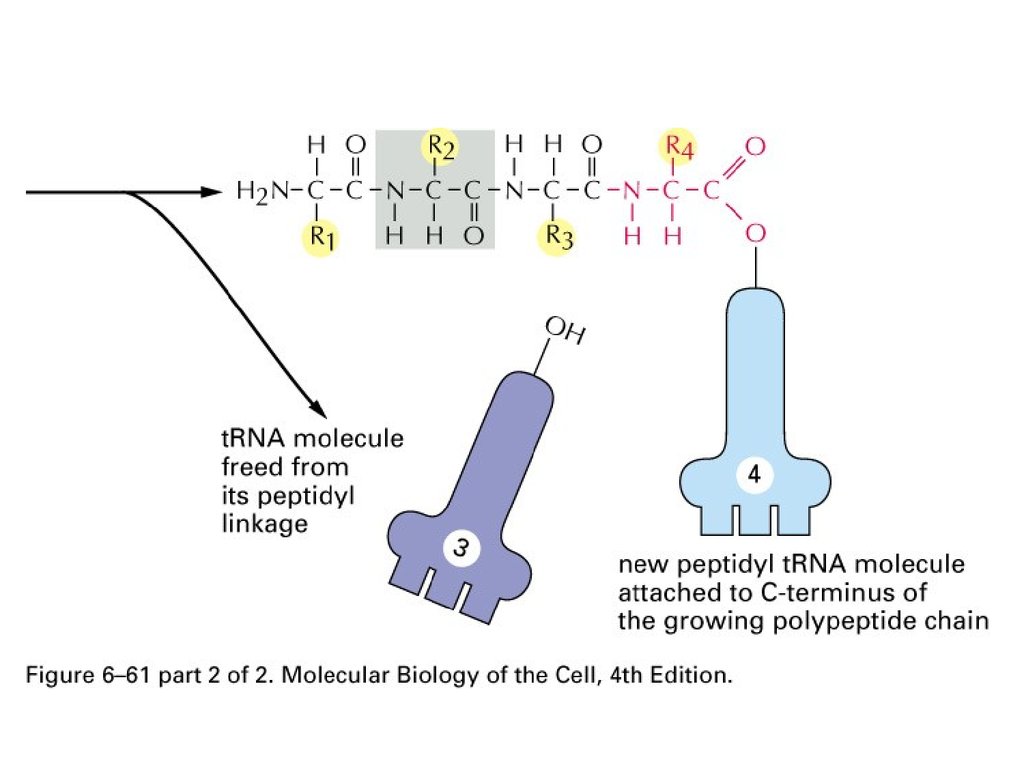
Protein synthesis65.
66.
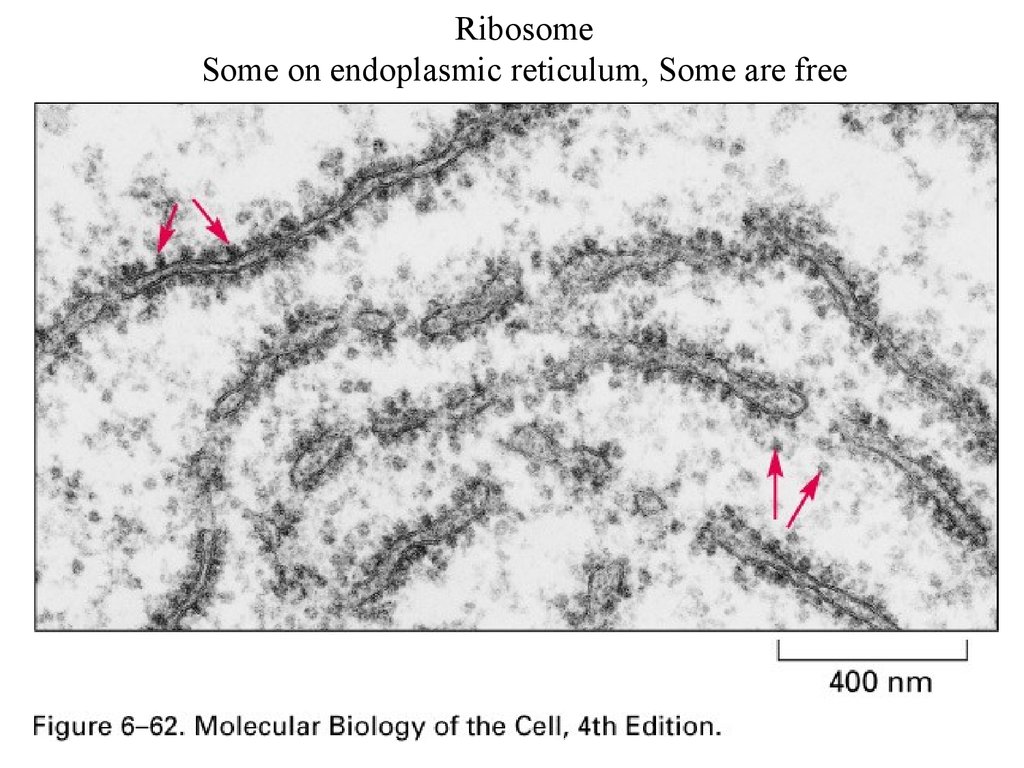
RibosomeSome on endoplasmic reticulum, Some are free
67.
Ribosome binding sites2 subunits: large and small
4 binding sites: 1 for mRNA at small subunit, 3 for tRNA in large subunit
68.
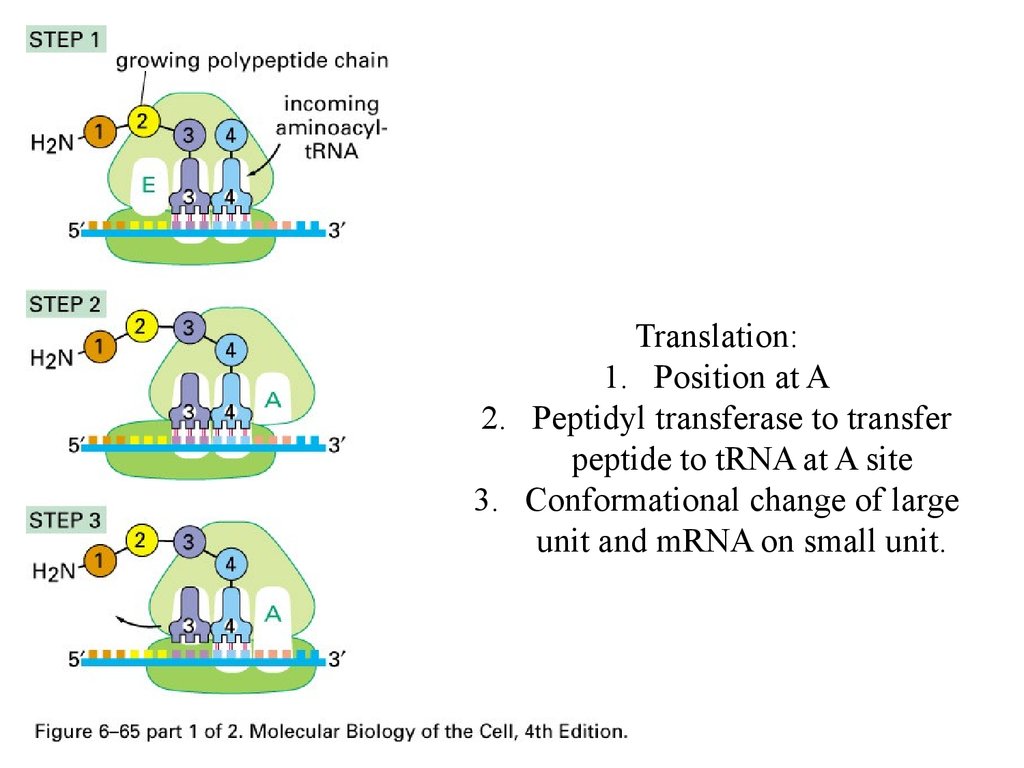
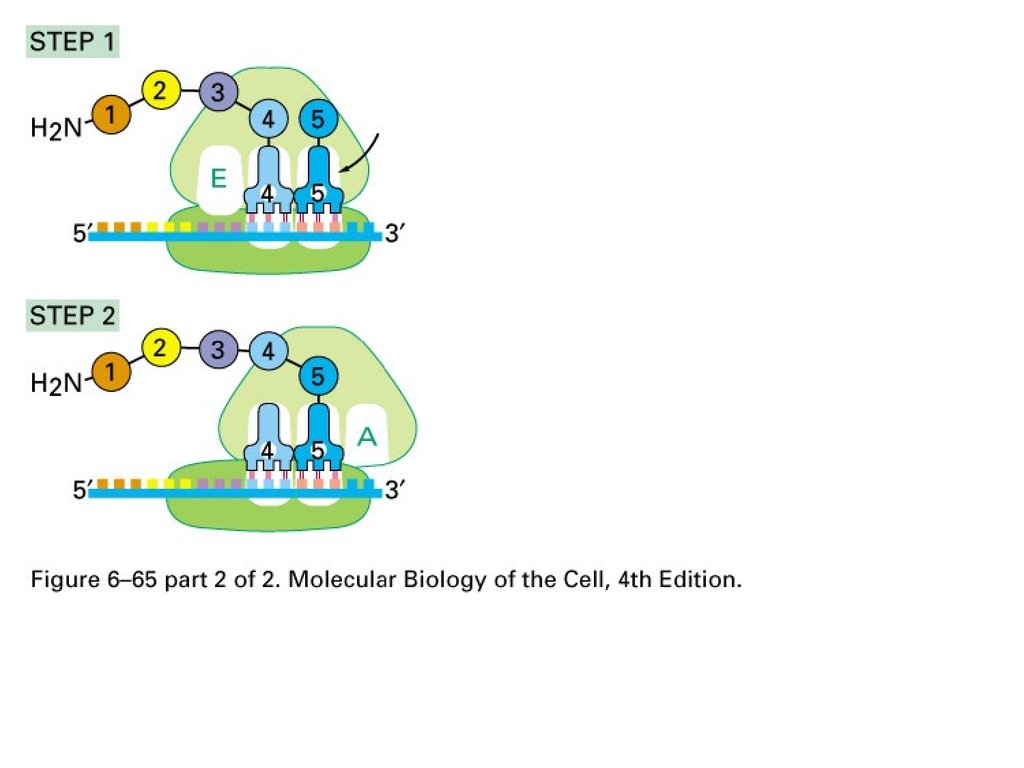
Translation:1. Position at A
2. Peptidyl transferase to transfer
peptide to tRNA at A site
3. Conformational change of large
unit and mRNA on small unit.
69.
70.
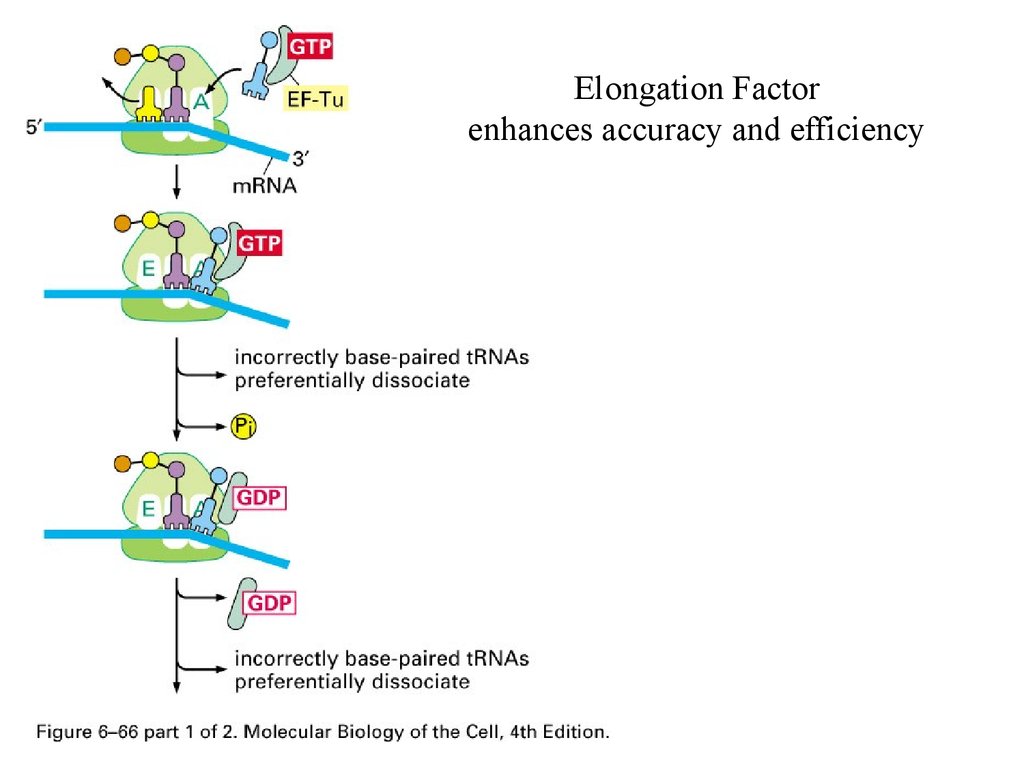
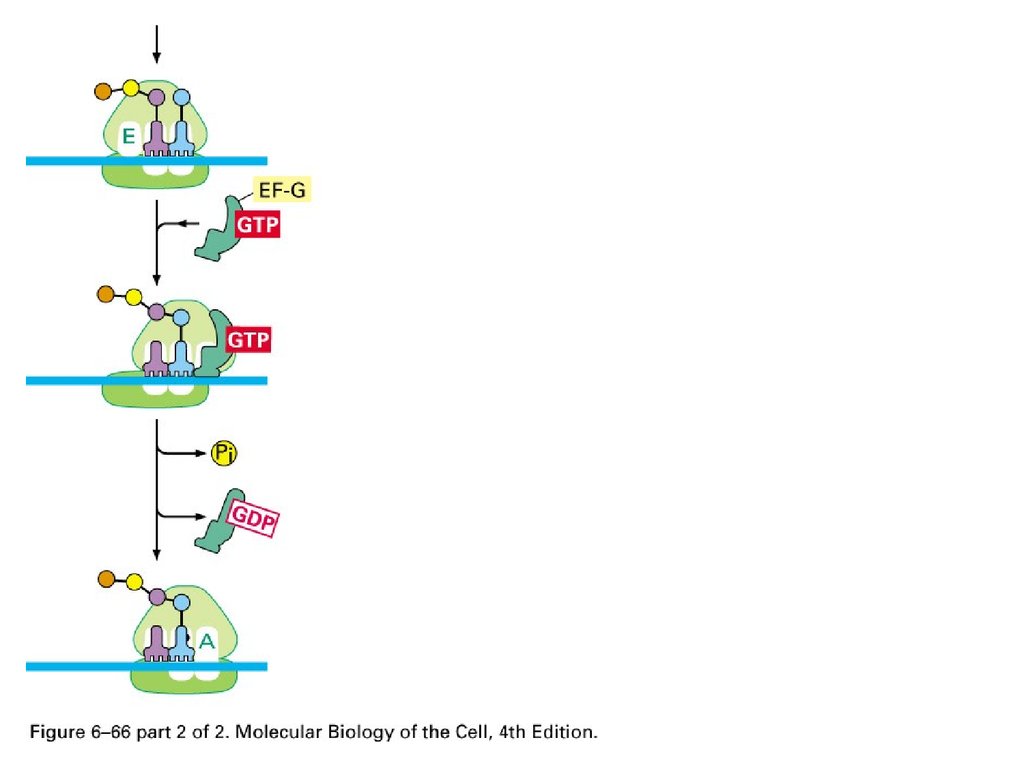
Elongation Factorenhances accuracy and efficiency
71.
72.
The Initiation of protein synthesis ineucaryotes
Eucaryotic initiation factors (eIFs)
AUG encodes Met
73.
74.
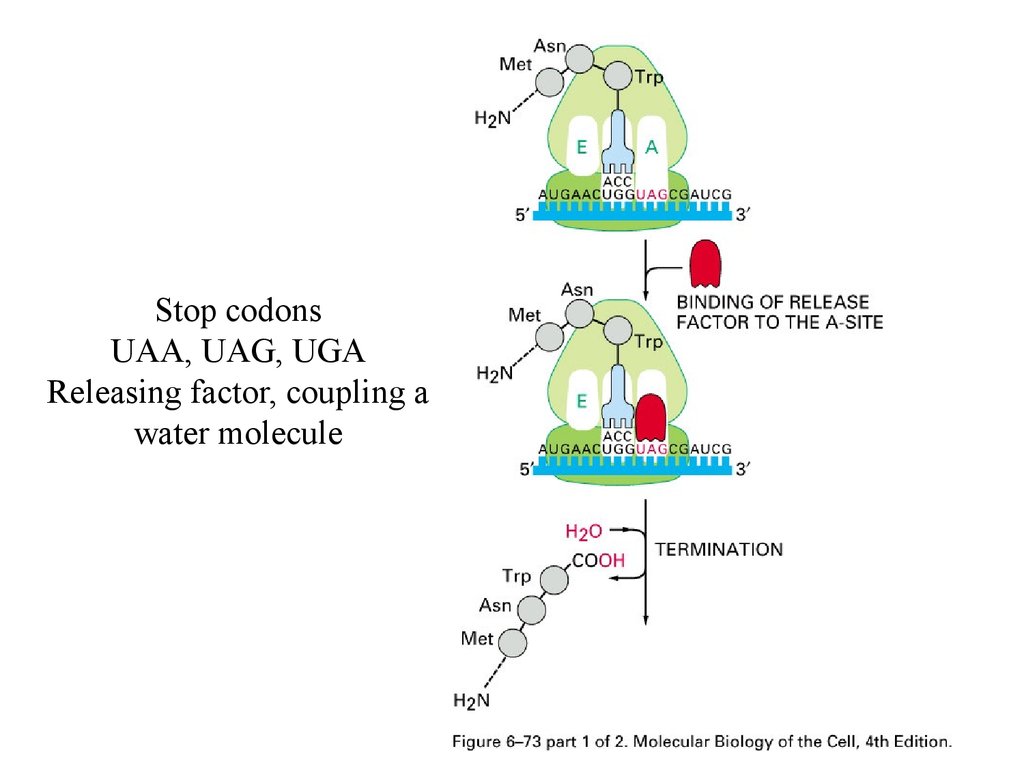
Stop codonsUAA, UAG, UGA
Releasing factor, coupling a
water molecule
75.
76.
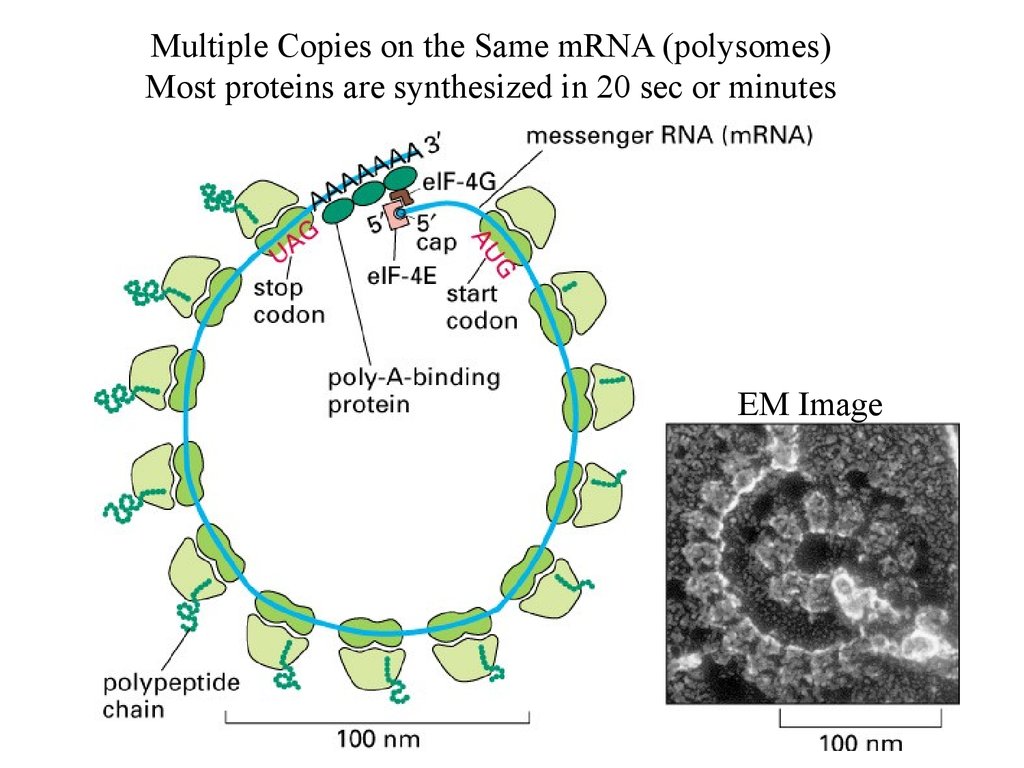
Multiple Copies on the Same mRNA (polysomes)Most proteins are synthesized in 20 sec or minutes
EM Image


























 biology
biology








