Similar presentations:
Introduction & overview
1.
PROTEIN PHYSICS1. Introduction & overview
2. Structure elements & elementary interactions
3. Transitions: thermodynamics & kinetics
4. Secondary structures
5. Protein structures
6. Protein denaturation & folding
7. Protein structure prediction, engineering, design
8. Proteins in action
2.
Олег Борисович Птицын(1929-1999)
3.
PROTEIN PHYSICSLECTURE 1
Introduction & overview
4.
Globularproteins
Membrane
proteins
Fibrous proteins
H-bonds (NH:::OC) & hydrophobic forces
5.
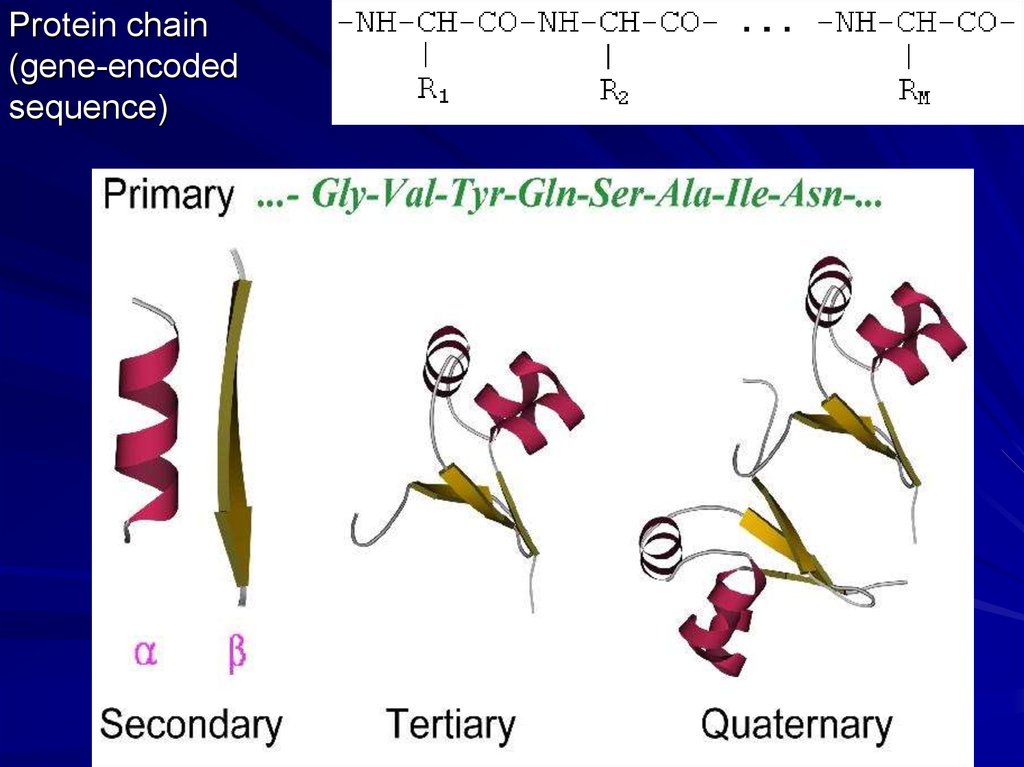
Protein chain(gene-encoded
sequence)
6.
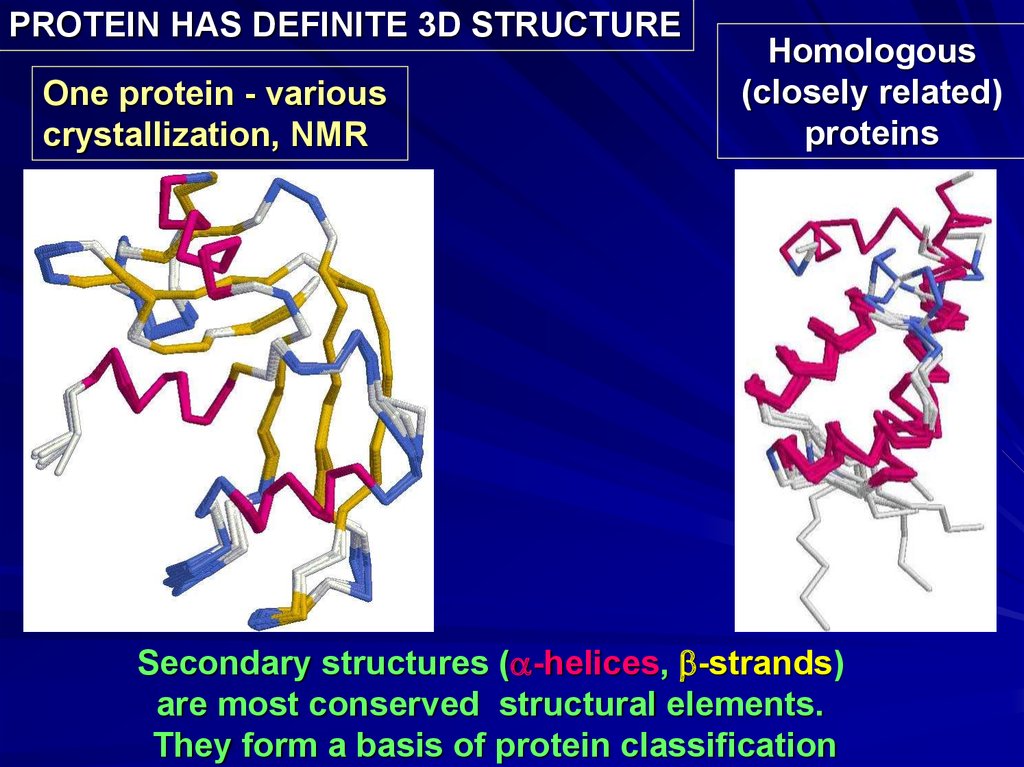
PROTEIN HAS DEFINITE 3D STRUCTUREOne protein - various
crystallization, NMR
Homologous
(closely related)
proteins
Secondary structures (a-helices, b-strands)
are most conserved structural elements.
They form a basis of protein classification
7.
8.
9.
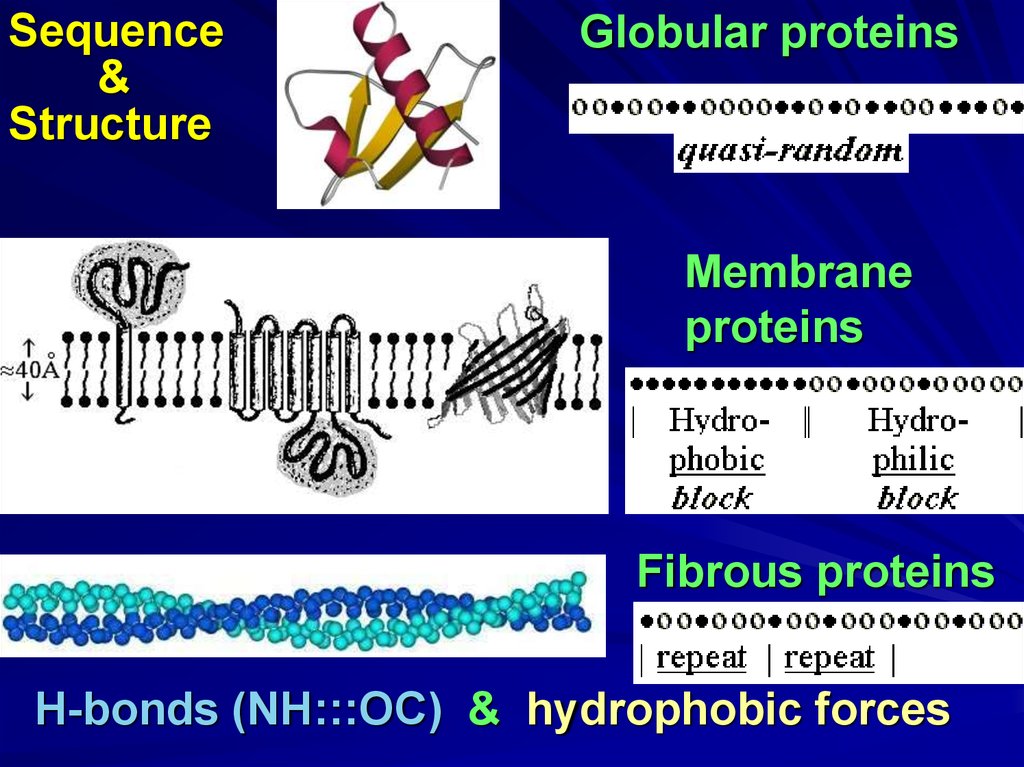
Sequence&
Structure
Globular proteins
Membrane
proteins
Fibrous proteins
H-bonds (NH:::OC) & hydrophobic forces
10.
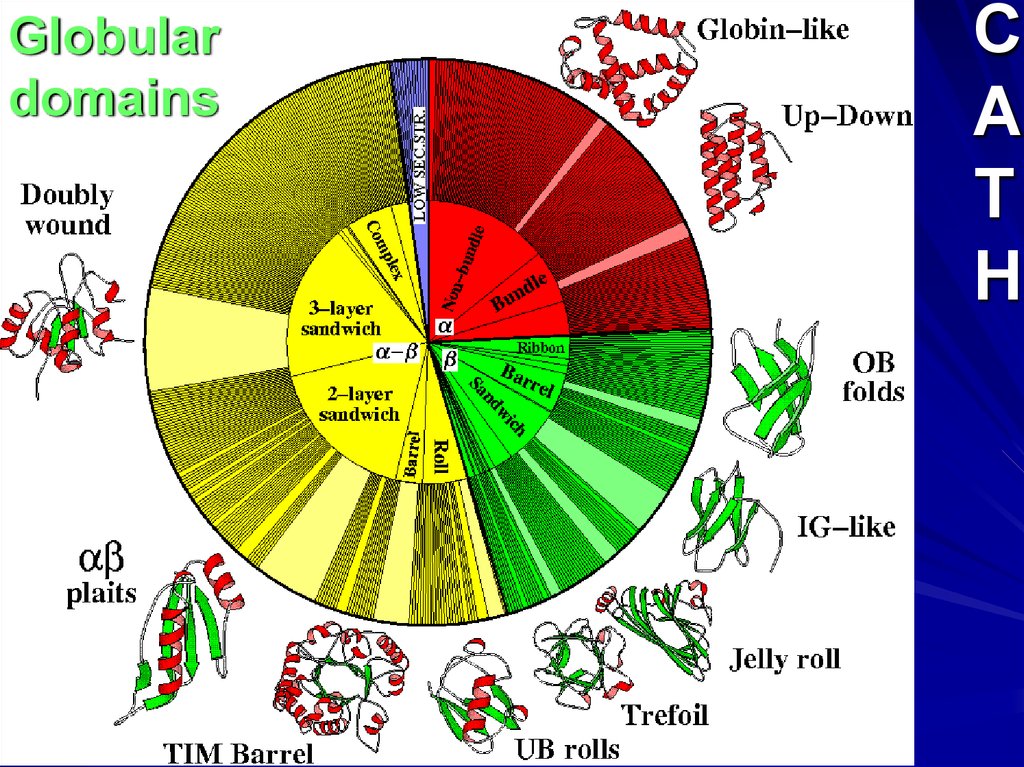
Globulardomains
C
A
T
H
11.
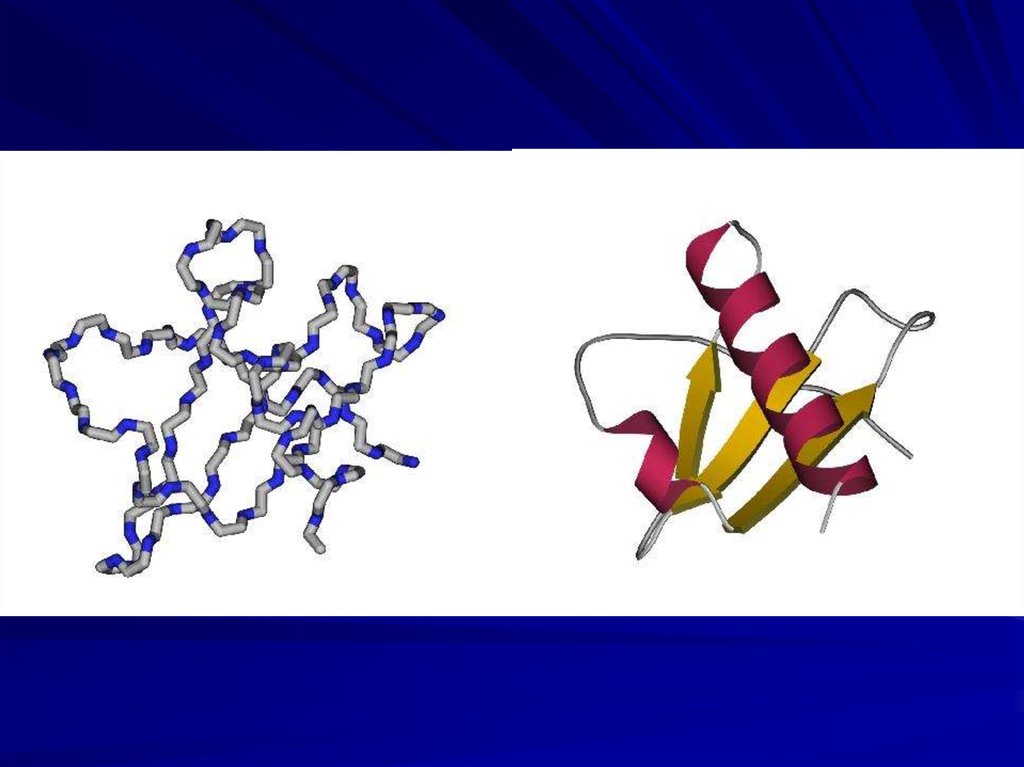
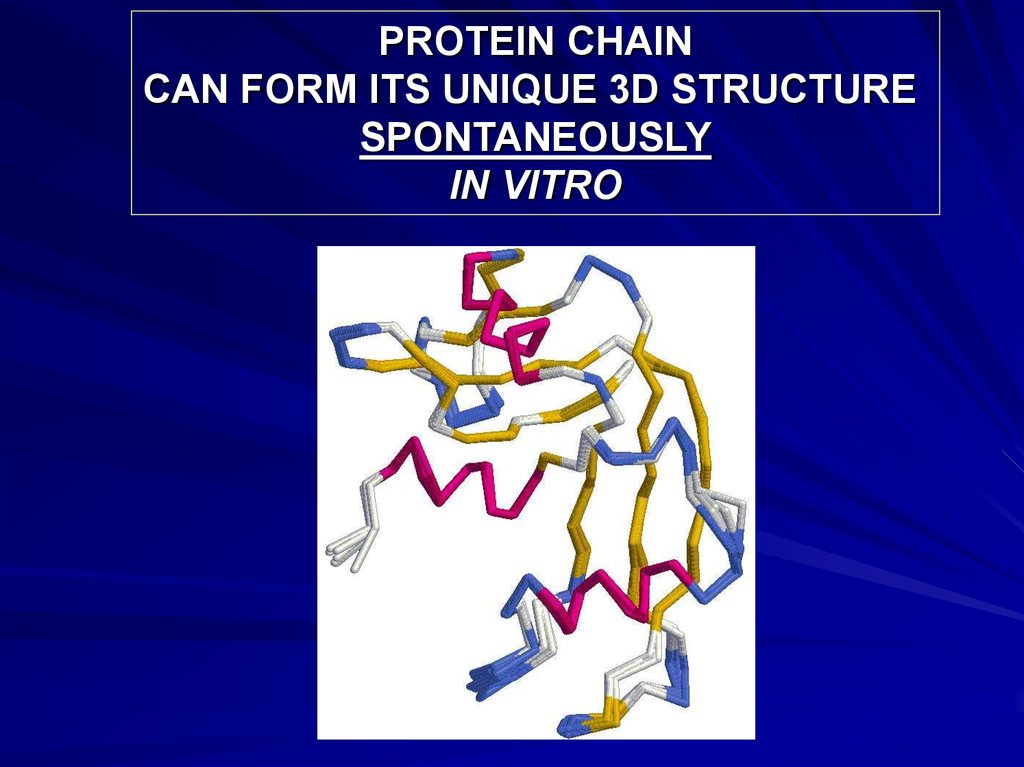
PROTEIN CHAINCAN FORM ITS UNIQUE 3D STRUCTURE
SPONTANEOUSLY
IN VITRO
12.
phase separation13.
BIND TRANSFORM RELEASE:ENZYMES (chymotrypsin)
Note small active site
14.
POST-TRANSLATIONAL MODIFICATIONSSometimes,
CHAIN CUT-INDUCED DEFORMATION
MAKES ENZYME ACTIVE
active
cat. site
Chymotripsin
nonactive
cat. site
Chymotripsinogen
15. POST-TRANSLATIONAL MODIFICATIONS: (especially in eukaryotes): PROTEIN CHAIN CUTS (proteolysis), - SPLICING (inteins) -
CYCLIZATION- INTERNAL CHEM. TRANSFORMATION
GLYCOSYLATION, etc.
MODIFICATION OF ENDS (acetylation, etc.)
MODIFICATION OF SIDE CHAINS (S-S bonding,
phosphorilation, etc.)
COFACTORS …
16.
Sometimes:Different folds with the same active site:
the same biochemical function
17.
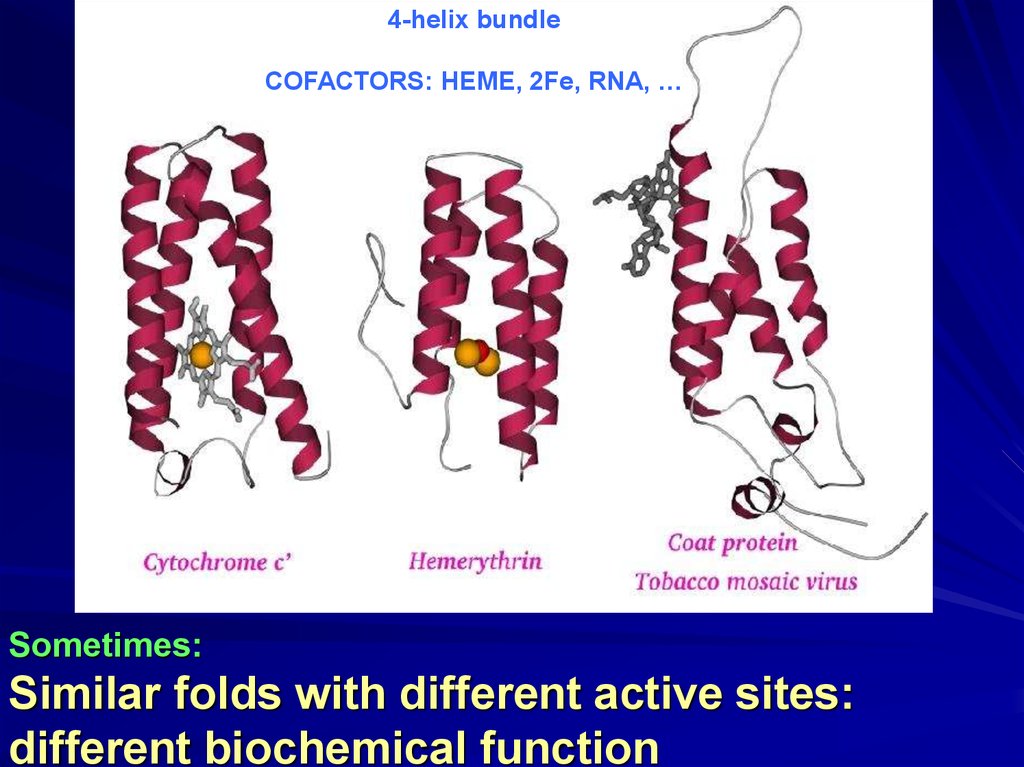
4-helix bundleCOFACTORS: HEME, 2Fe, RNA, …
Sometimes:
Similar folds with different active sites:
different biochemical function
18.
Standard positions of active sitesin protein folds
19.
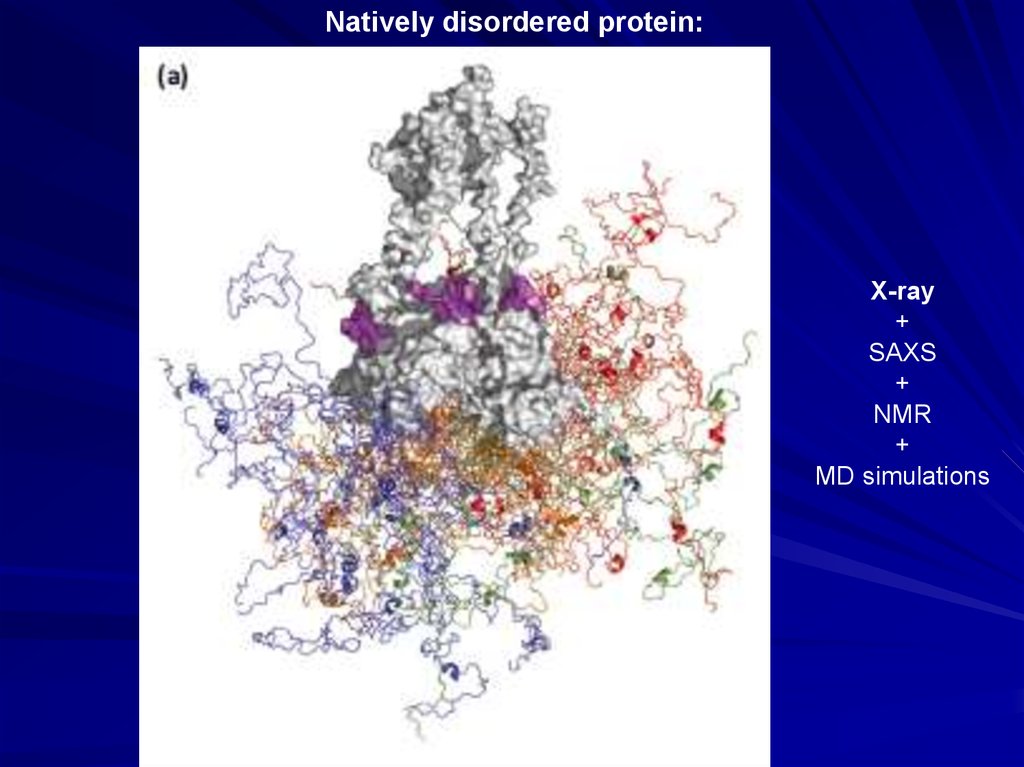
Natively disordered protein:X-ray
+
SAXS
+
NMR
+
MD simulations
20.
Chaperone GroEL21.
NMR______
22.
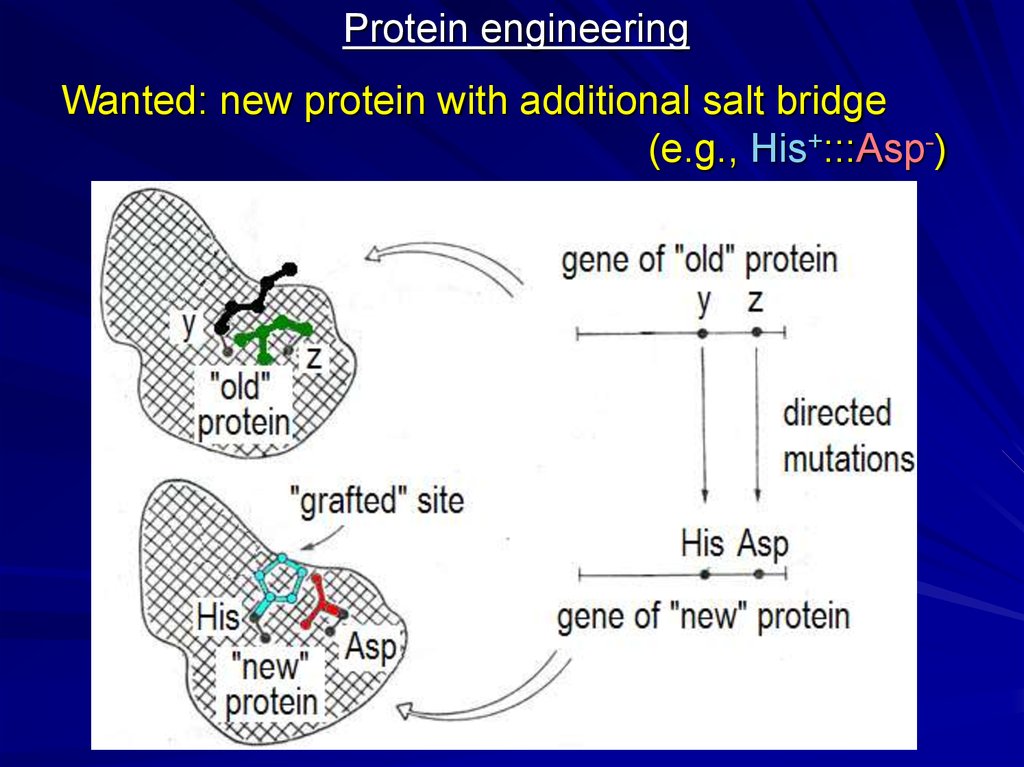
Protein engineeringWanted: new protein with additional salt bridge
(e.g., His+:::Asp-)
23.
PROTEIN PHYSICSLECTURE 2
Elementary interactions:
covalent
24.
Protein chain:regular backbone
&
gene-encoded sequence
of side chains
25.
Protein chainCovalent bond
lengths:
0.9 – 1.8 Å
Covalent bond
angles:
109o – 120o
Atom radii:
1–2Å
26.
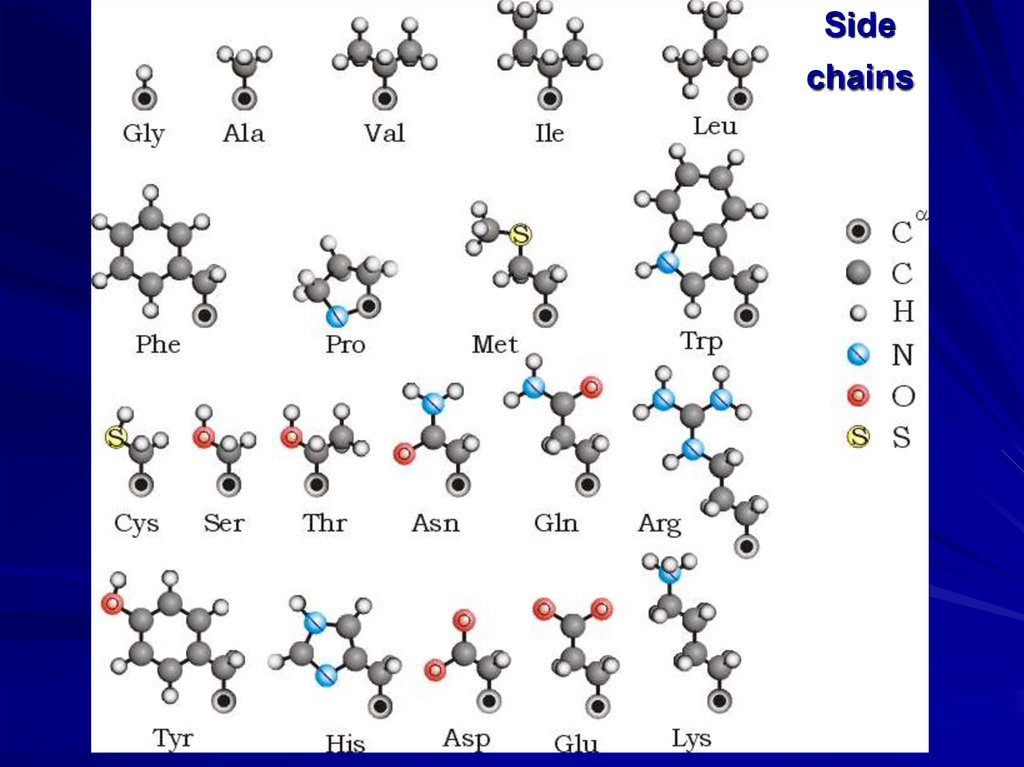
Sidechains
27.
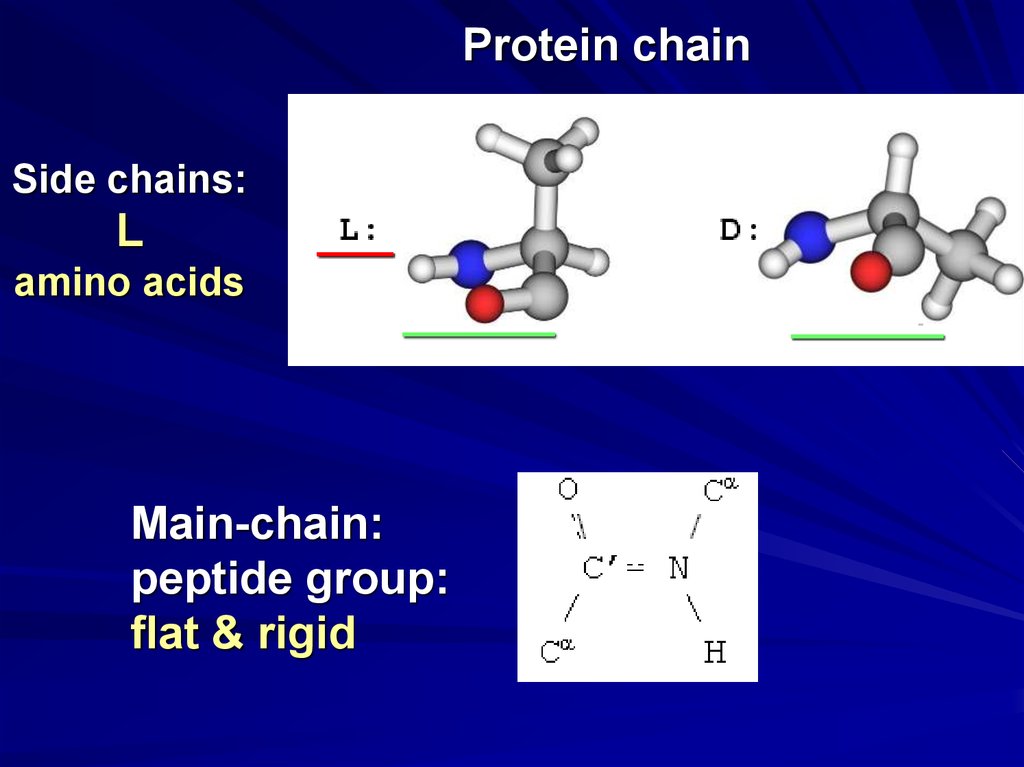
Protein chainSide chains:
L
amino acids
___
______
Main-chain:
peptide group:
flat & rigid
______
28.
Stereo imagesSymmetric
Asymmetric
backbone-toside_chain:
Two
asymmetric
side
chains:
Gly
Ala _L
Thr
Ile
29.
V = ±|V|semi-classical
approximation
~
30.
Werner Karl Heisenberg (1901-76)— Nobel Prize 1932
Wolfgang Ernst Pauli ) (1900-58)
— Nobel Prize 1945
31.
Peptide group:flat & rigid
Pauling resonance
theory of = bonds:
O=C-N ↔ O-C=N
O C N
Linus Carl
Pauling
(1901-94)
— Nobel Prizes:
1954, 62
Covalent bonding in peptide group:
sp2 + p
O
sp2 + p
O
=
32.
Main-chain:f (N-Ca) ,
y (Ca-C’),
w (C’=N)
Side-chain:
c1, c2, ...
33.
Countingangles:
120o
180o
0o
_____________________________________________
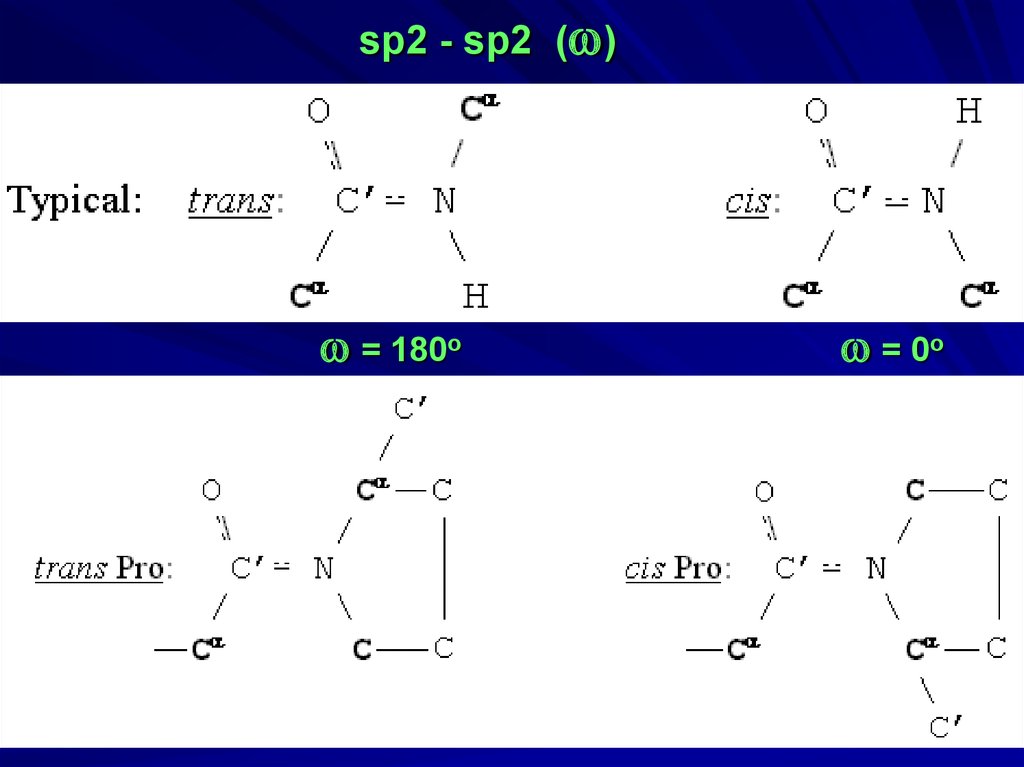
34.
sp2 - sp2 (w)w = 180o
w = 0o
35.
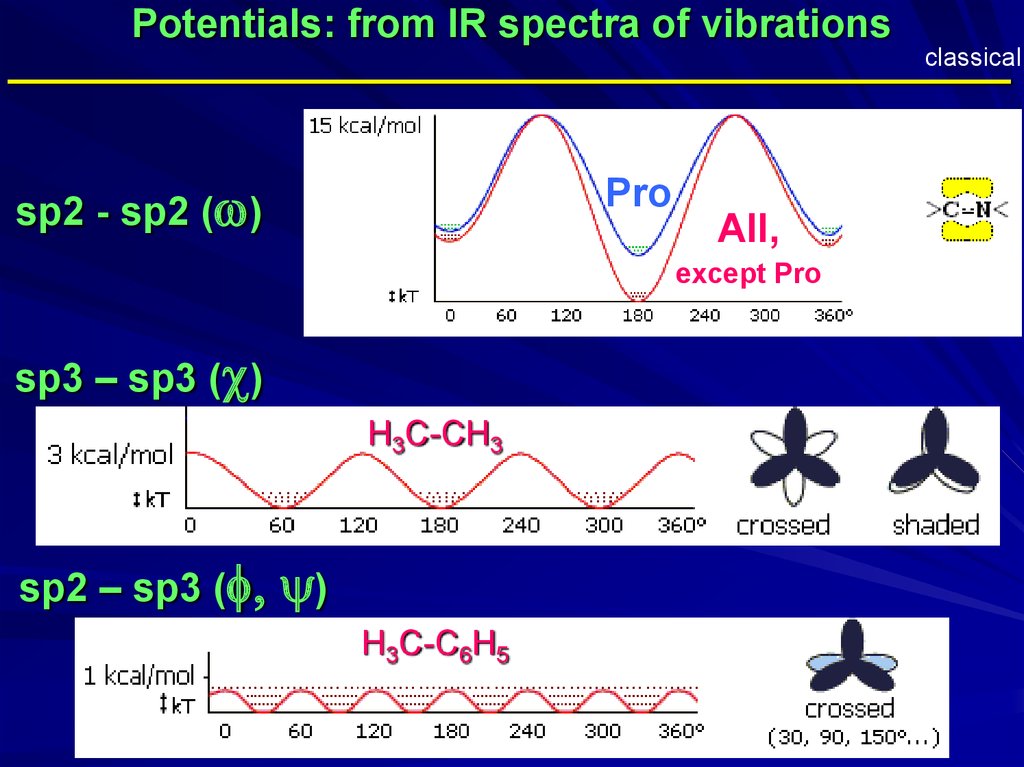
Potentials: from IR spectra of vibrationsclassical
_____________________________________________
sp2 - sp2 (w)
Pro
All,
except Pro
sp3 – sp3 (c)
H3C-CH3
sp2 – sp3 (f,
y)
H3C-C6H5
36.
Поворотно-изомерная теория полимеровМихаил Владимирович
Волькенштейн (1912-92)
Олег Борисович
Птицын (1929-99)
Paul John Flory (1910-85)
— Nobel Prize 1974
Конформационный анализ
Александр
Исаакович
Китайгородский
(1914–1985)
Harold
Abraham
Scheraga
(1921)
37.
The Nobel Prize in Chemistry 2013Martin Karplus
Michael Levitt
Arieh Warshel
"for the development of multiscale models
for complex chemical systems"
(conformational & quantum-mechanical methods)























 physics
physics








