Similar presentations:
Structures of water-soluble globular proteins
1.
PROTEIN PHYSICSLECTURE 13-16
- Structures of water-soluble globular proteins
- Physical selection of protein structures
- Structural classification of proteins
2.
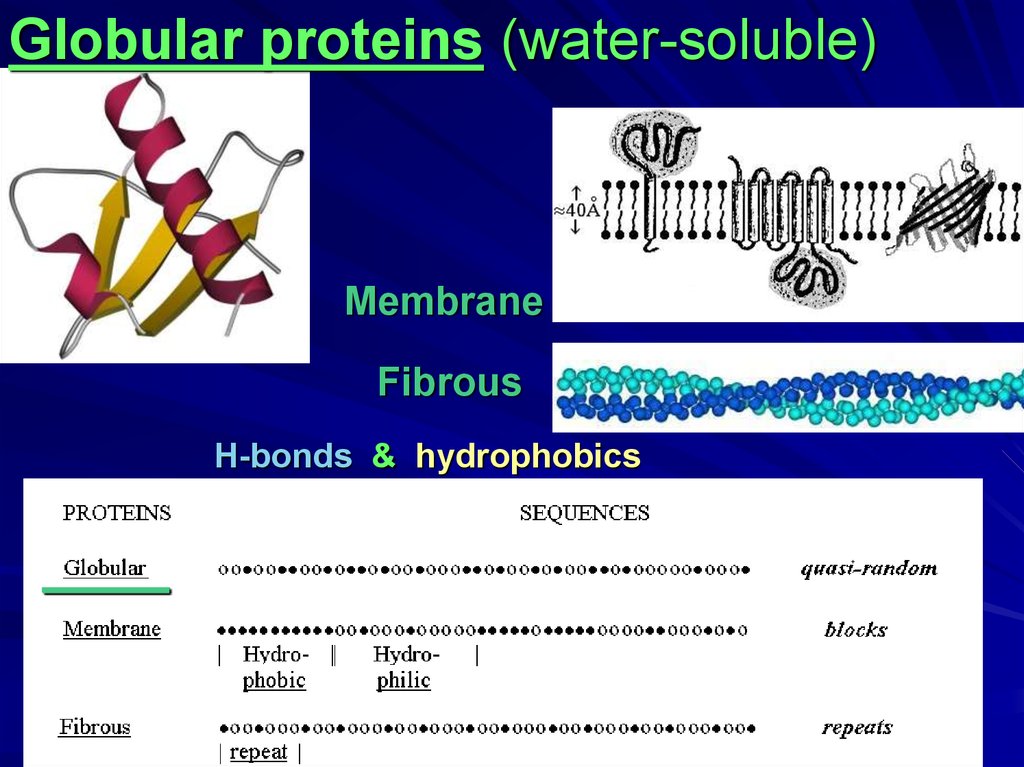
Globular proteins (water-soluble)Membrane
Fibrous
H-bonds & hydrophobics
____
3.
Protein chainHermann Emil Louis
Fischer
(1852 –1919)
Nobel Prize 1902
Protein sequence
Frederick Sanger
(1918 –2013)
Nobel Prizes: 1958, 1980
4.
single-domainglobular protein
domain 1 domain 2
fold
stack
5.
X-RAYOne protein, various
crystallizations
NMR
Homologous
Structures, compatible
with one NMR experiment
(closely related)
proteins
Secondary structures (a-helices, b-strands) are
the most rigid and conserved details of proteins;
they are determined with the smallest errors and
form a basis of protein classification
6.
X-ray 3D protein structureMax Ferdinand Perutz
(1914 –2002)
Nobel Prize 1962
NMR 3D protein structure
Kurt Wüthrich, 1938
Nobel Prize 2002
7.
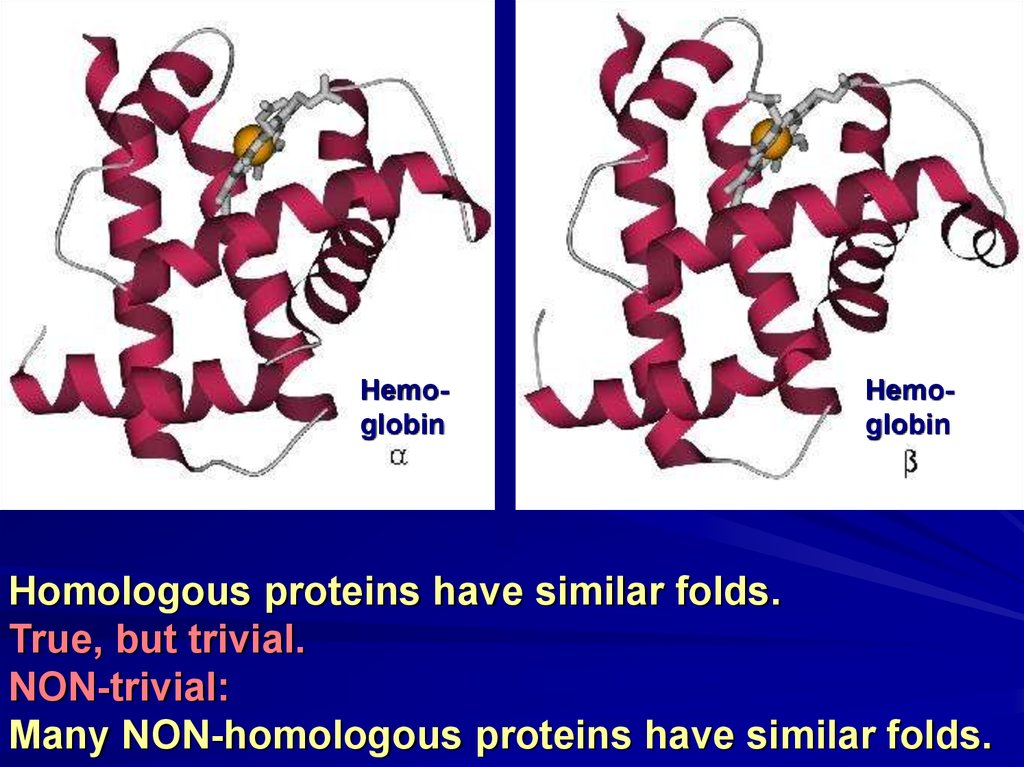
HemoglobinHemoglobin
Homologous proteins have similar folds.
True, but trivial.
NON-trivial:
Many NON-homologous proteins have similar folds.
8.
b-sheets: usually, twisted(usually, right-)
____
b-proteins
H-bonds: within sheets
Hydrophobics: between sheets
9.
sandwiches&
cylinders
Orthogonal packing
of b-sheets
Aligned packing
of b-sheets
10.
Retinol-binding proteinorthogonal packing
of one rolled b-sheet
11.
55
2’
3
6 4 5’
4
2’
6
3
1
1
2
2
Trypsin-like SER-protease
Acid-protease
orthogonal packings of b-sheets
5’
12.
54 3
2
6
7
Greek key 2::5
Greek key 3::6
non-crossed loops
1
IG-fold:
aligned packing of b-sheets
13.
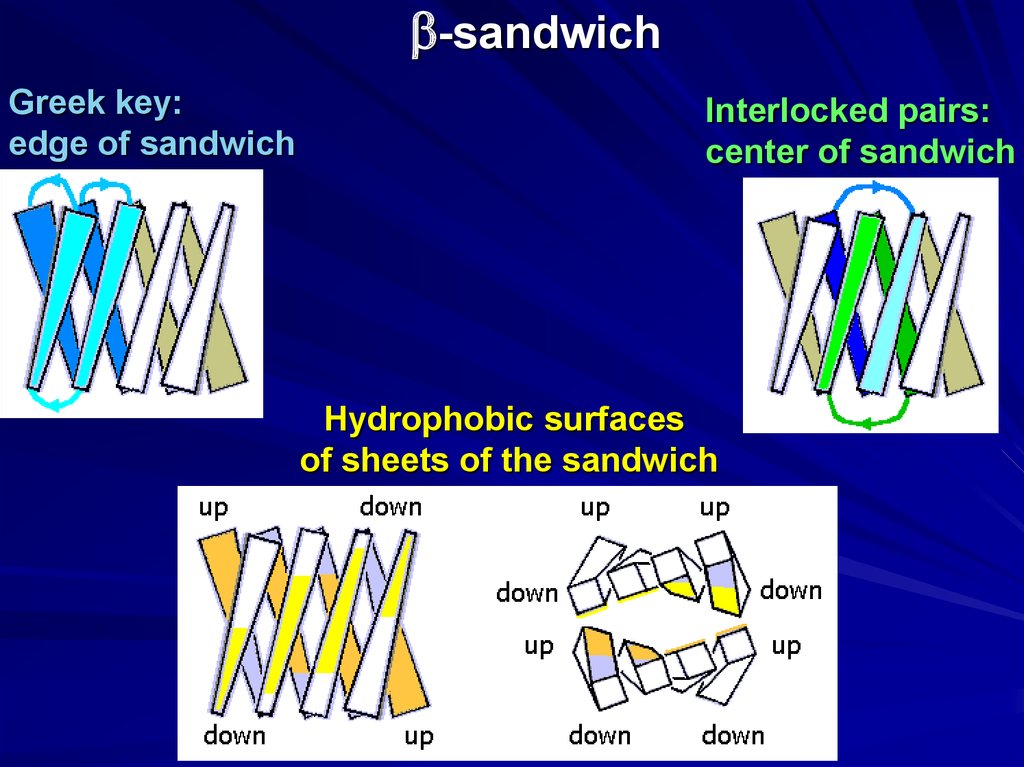
b-sandwichGreek key:
edge of sandwich
Interlocked pairs:
center of sandwich
Hydrophobic surfaces
of sheets of the sandwich
14.
16 5
8
2
6
2
3
3
8 1
6
1
3
8
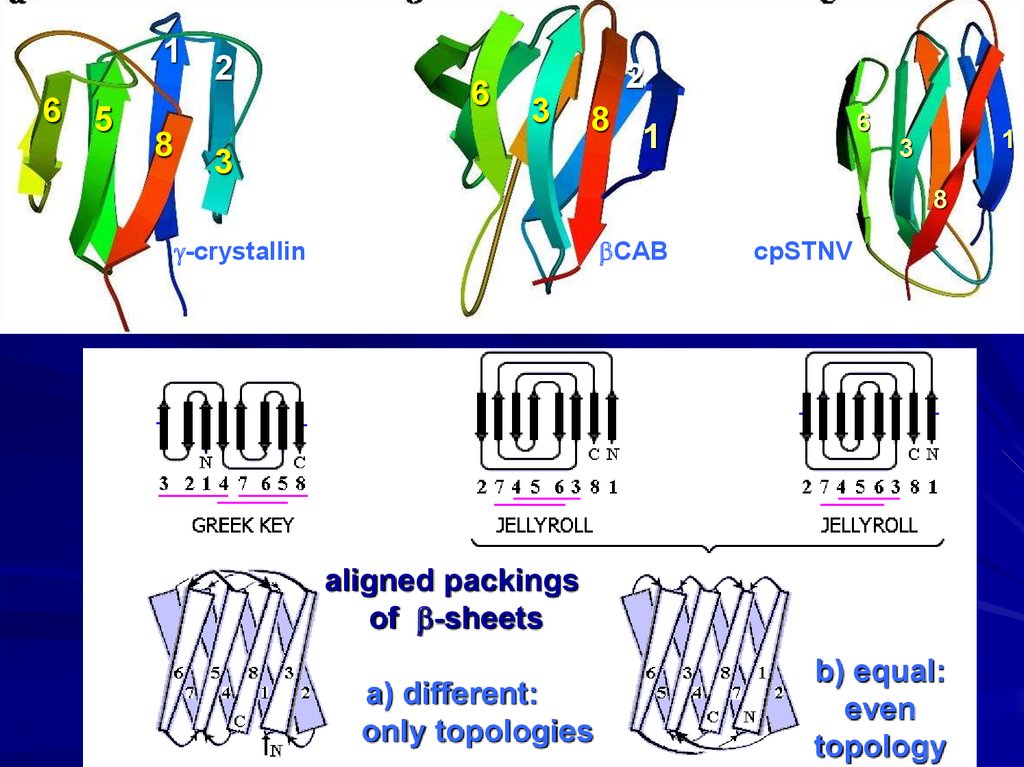
g-crystallin
bCAB
cpSTNV
aligned packings
of b-sheets
a) different:
only topologies
b) equal:
even
topology
15.
aligned packingof b-sheets
6-bladed propeller
neuraminidase
16.
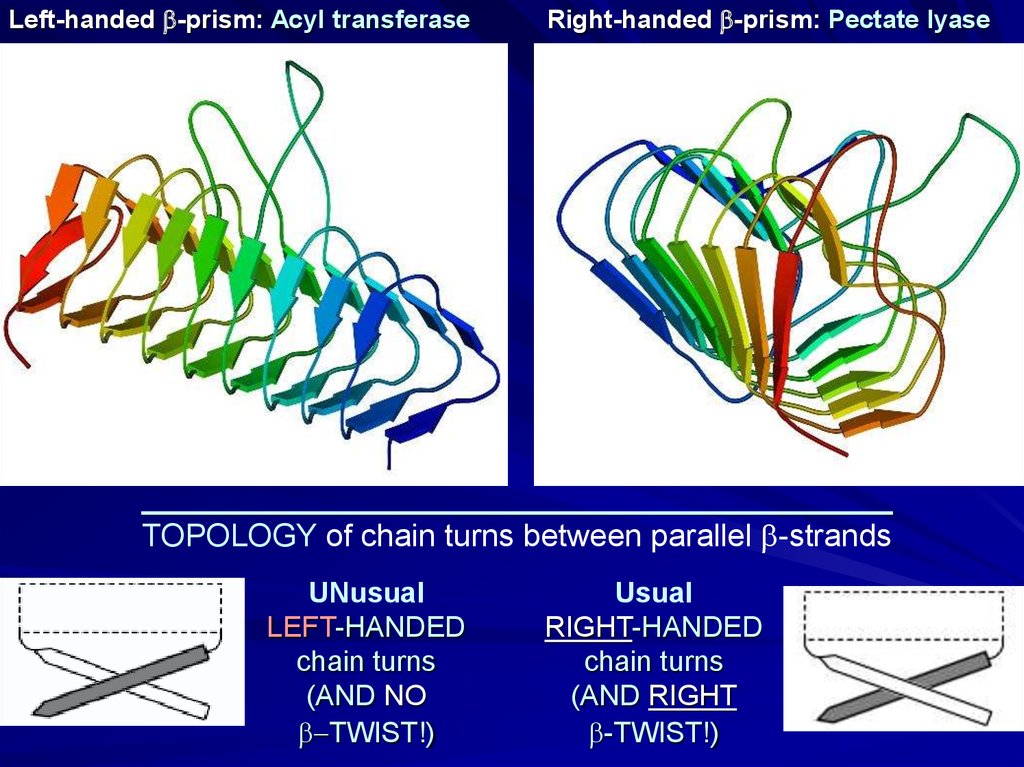
Left-handed b-prism: Acyl transferaseRight-handed b-prism: Pectate lyase
___________________________________________
TOPOLOGY of chain turns between parallel b-strands
UNusual
LEFT-HANDED
chain turns
(AND NO
b-TWIST!)
Usual
RIGHT-HANDED
chain turns
(AND RIGHT
b-TWIST!)
17.
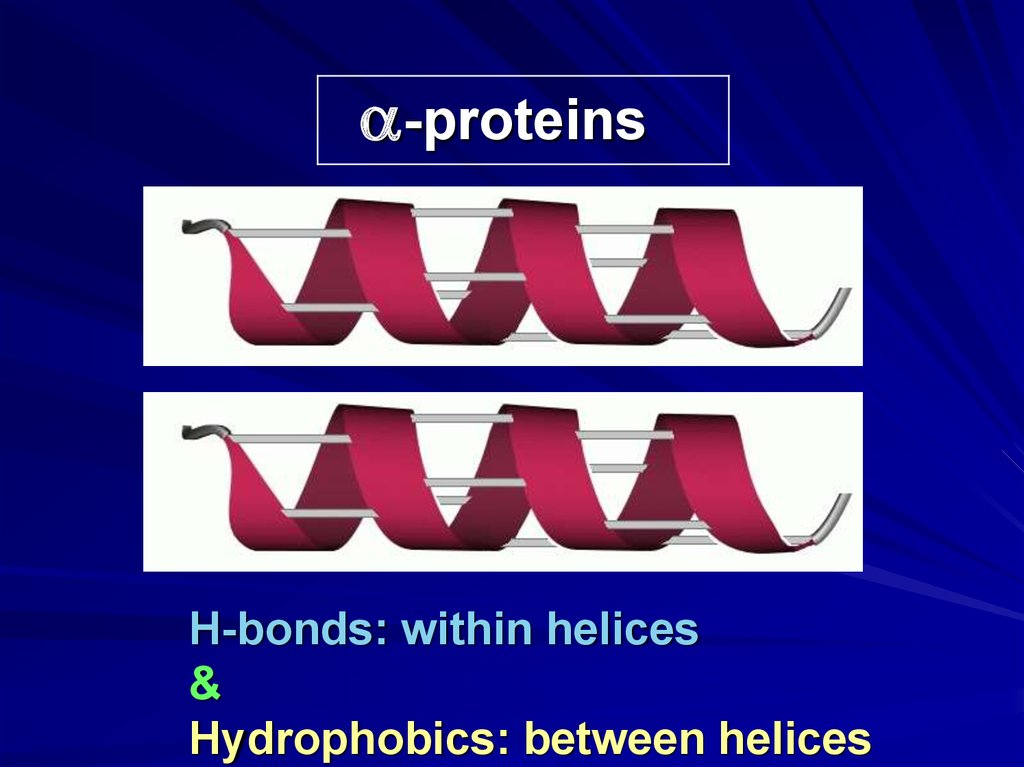
a-proteinsH-bonds: within helices
&
Hydrophobics: between helices
18.
Quasi-cylindrical core (in fibrous)Quasi-flat core
Quasi-spherical core
MOST COMMON
19.
Orthogonal packingof LONG a-helices
Similar to orthogonal
packing of b-sheets
20.
Aligned packingof LONG a-helices
Similar to aligned
packing of b-sheets
21.
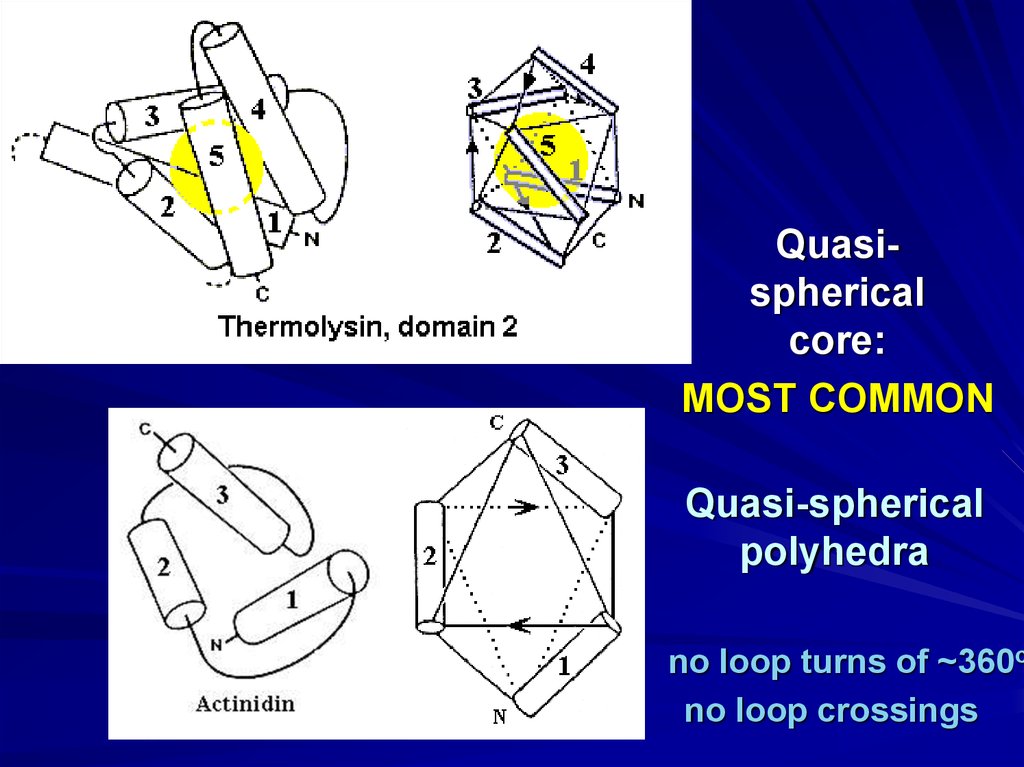
Quasisphericalcore:
MOST COMMON
Quasi-spherical
polyhedra
no loop turns of ~360o
no loop crossings
22.
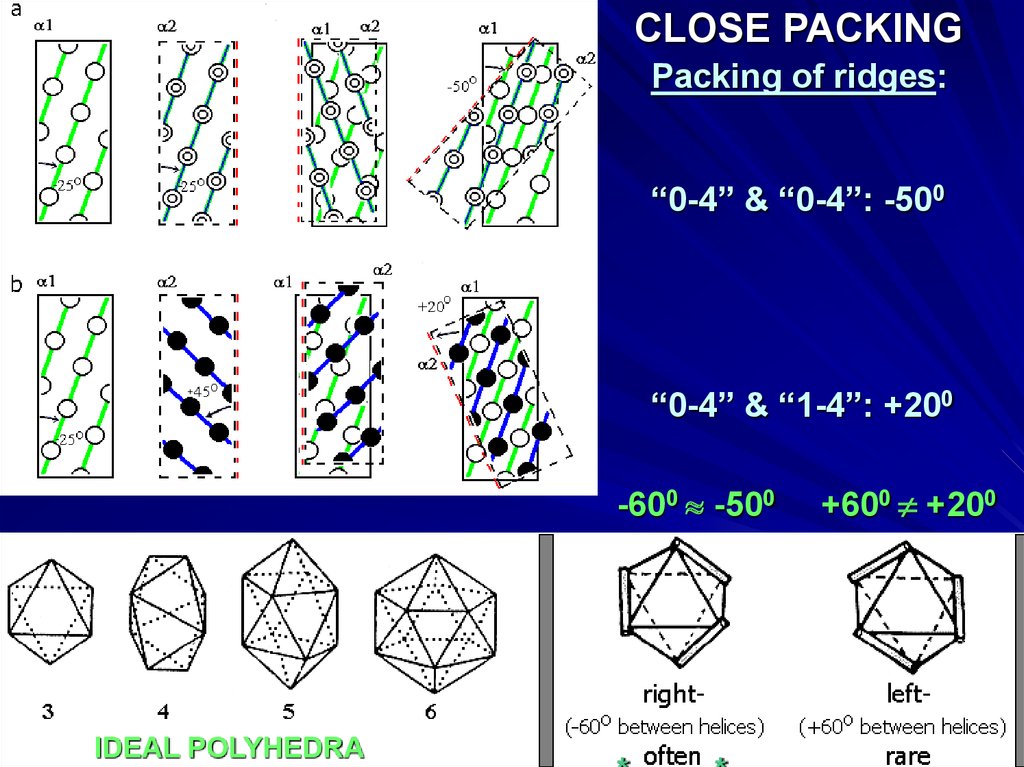
CLOSE PACKINGPacking of ridges:
“0-4” & “0-4”: -500
“0-4” & “1-4”: +200
-600 -500
IDEAL POLYHEDRA
+600 +200
23.
a/b proteinsH-bonds: within helices & sheets
Hydrophobics: between helices & sheets
24.
TIM barrelRossmann fold
25.
Regular secondary structure sequence:b - a - b - a - b - a - b - a - b - ...
a and b layers
right-handed
superhelices
26.
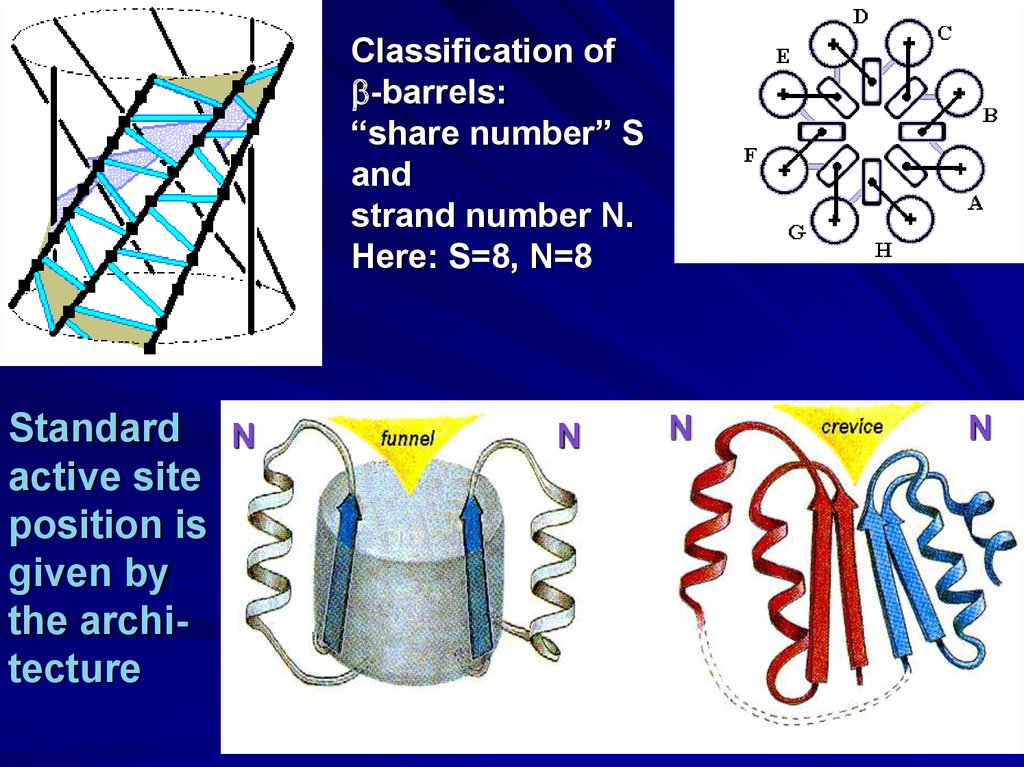
Classification ofb-barrels:
“share number” S
and
strand number N.
Here: S=8, N=8
Standard N
active site
position is
given by
the architecture
N
N
N
27.
a+b proteinsH-bonds: within helices & sheets
Hydrophobics: between helices & sheets
28.
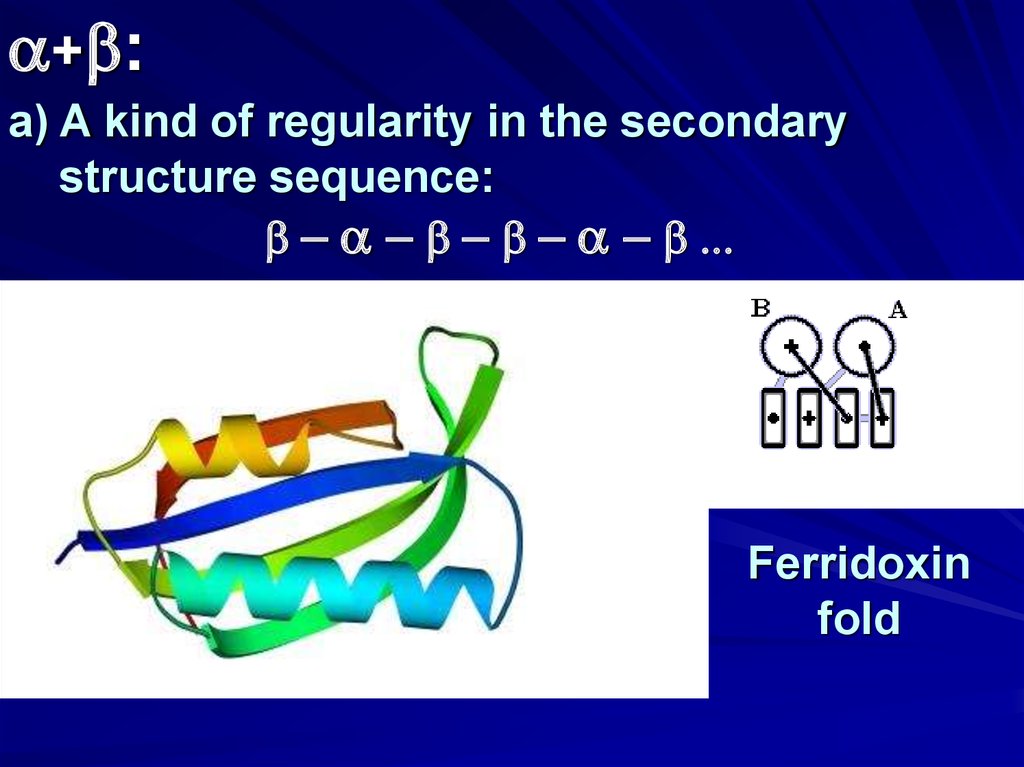
a+b:a) A kind of regularity in the secondary
structure sequence:
b - a - b - b - a - b ...
Ferridoxin
fold
29.
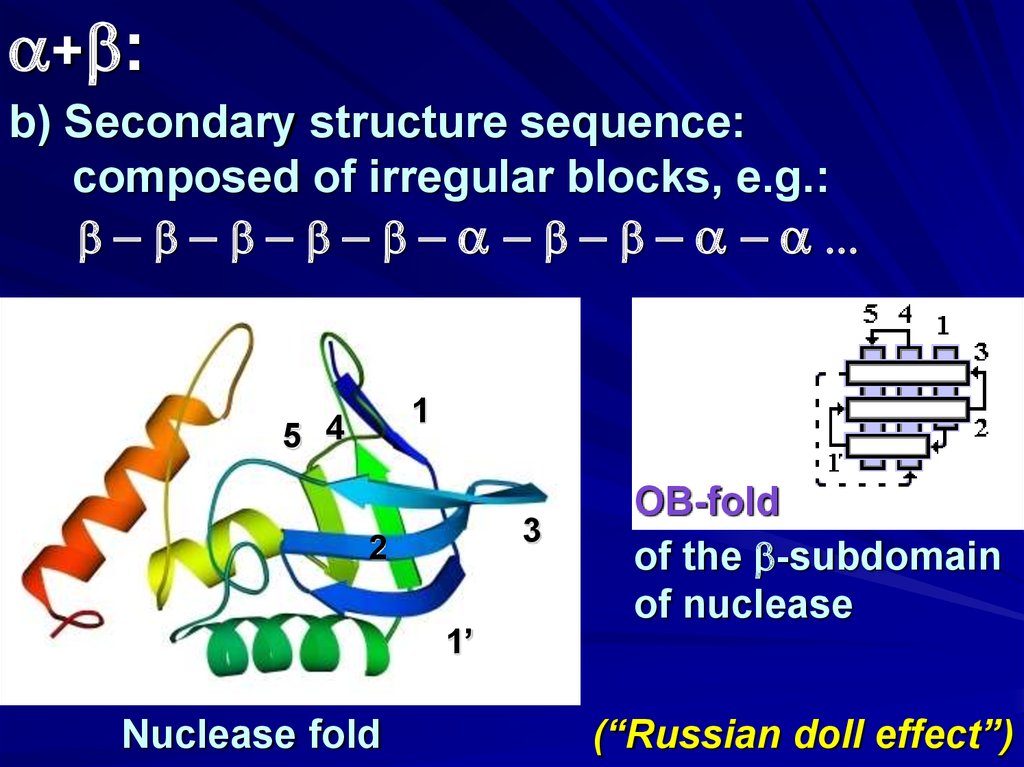
a+b:b) Secondary structure sequence:
composed of irregular blocks, e.g.:
b - b - b - b - b - a - b - b - a - a ...
1
5 4
3
2
1’
Nuclease fold
OB-fold
of the b-subdomain
of nuclease
(“Russian doll effect”)
30.
TYPICALFOLDING PATTERNS
(1977)
Jane Shelby
Richardson,
1941
31.
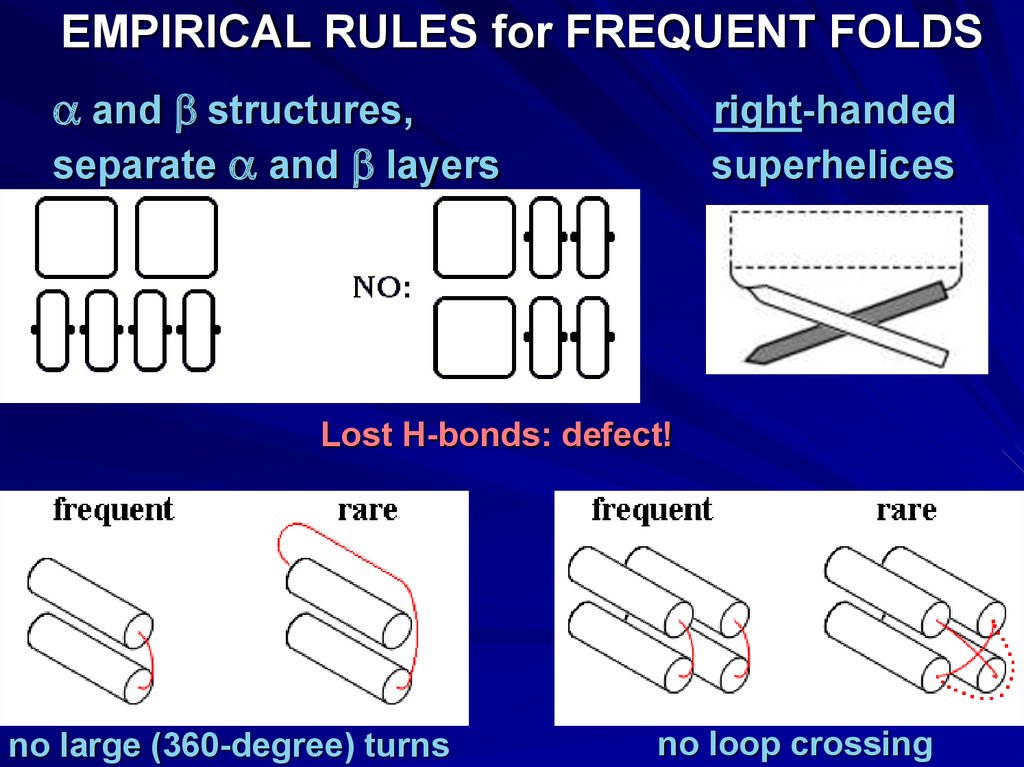
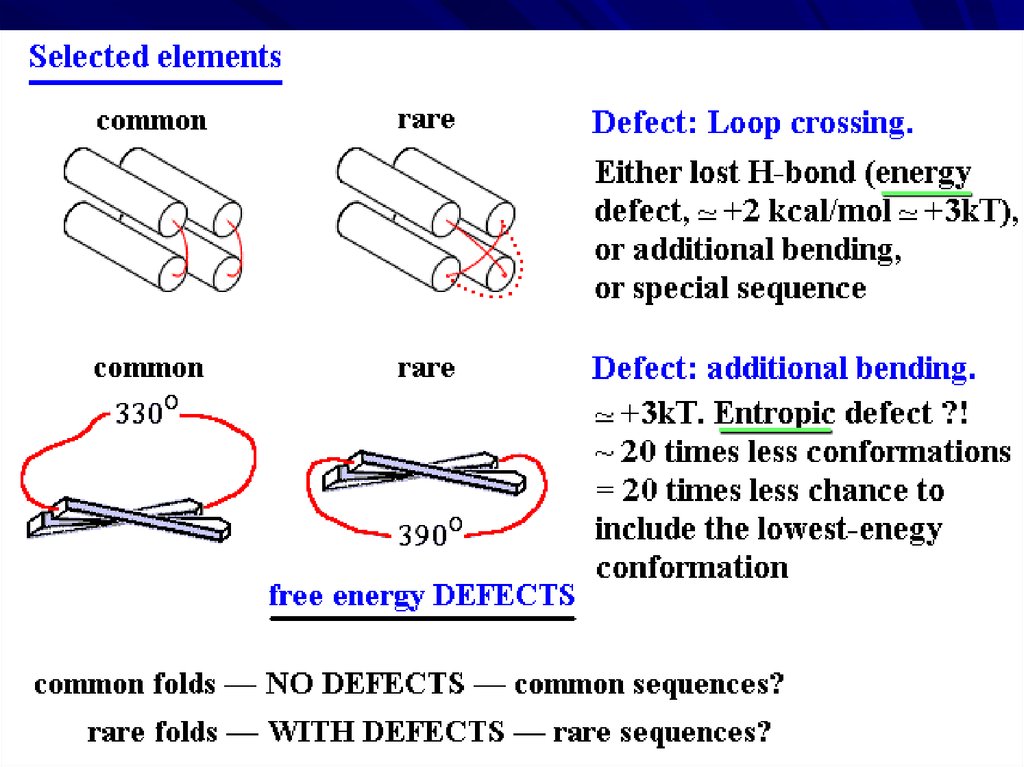
EMPIRICAL RULESseparate a and b layers
right-handed
superhelices
Lost H-bonds: defect!
no large, ~360o turns
NO ‘defects’
no loop crossings
32.
RESULT:NARROW SET
OF PREDOMINANT FOLDING PATTERNS
these are those that have no ‘defects’
33.
ALSO,these are “natively disordered proteins”,
which form a definite structure
only when bound
to some another molecule
(ligand, DNA, protein…)
34.
Globulardomains
C
A
T
H
S
C
O
P
35.
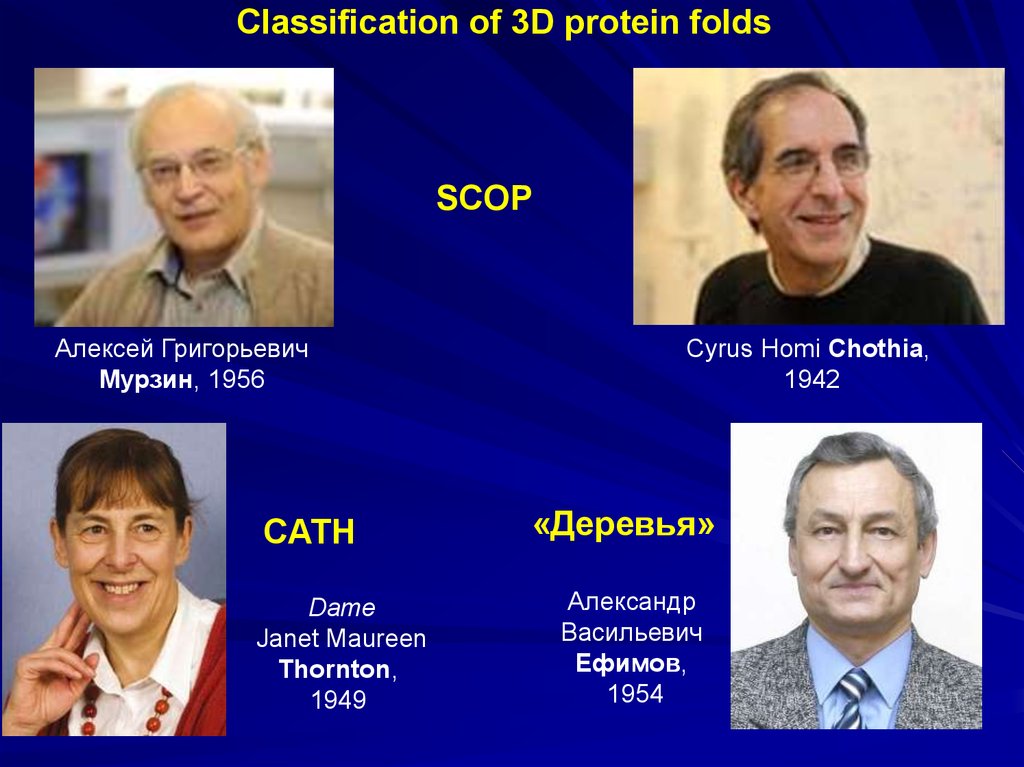
Classification of 3D protein foldsSCOP
Алексей Григорьевич
Мурзин, 1956
CATH
Dame
Janet Maureen
Thornton,
1949
Cyrus Homi Chothia,
1942
«Деревья»
Александр
Васильевич
Ефимов,
1954
36.
Efimov’s “trees”37.
80/20 LAW:38.
EMPIRICAL RULES for FREQUENT FOLDSa and b structures,
separate a and b layers
right-handed
superhelices
Lost H-bonds: defect!
no large (360-degree) turns
no loop crossing
39.
e.g.:Unusual fold
(no
a, almost no b structure: bad for stability) BUT: very special sequence
(very many Cysteins, and therefore
very many S-S bonds)
40.
Unusualfold (GFP):
helix inside
Usual folds:
helices outside
41.
What is more usual:sequence providing a inside or b b inside?
a
bb
N>150
42.
_________
43.
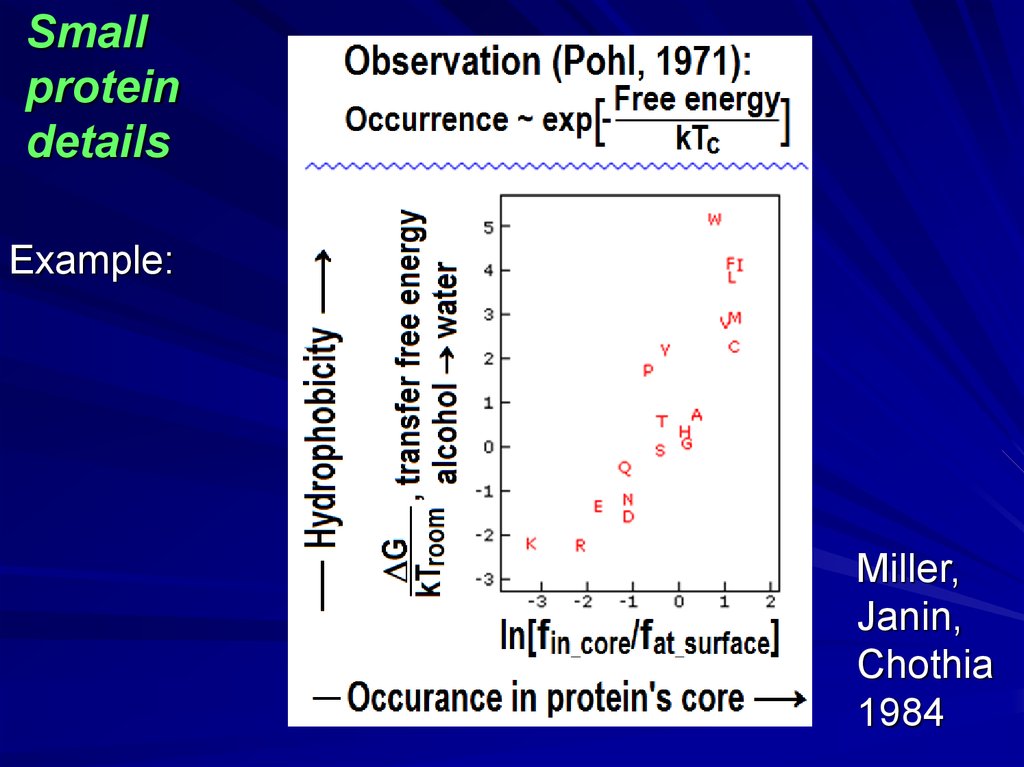
Smallprotein
details
Example:
Miller,
Janin,
Chothia
1984
44.
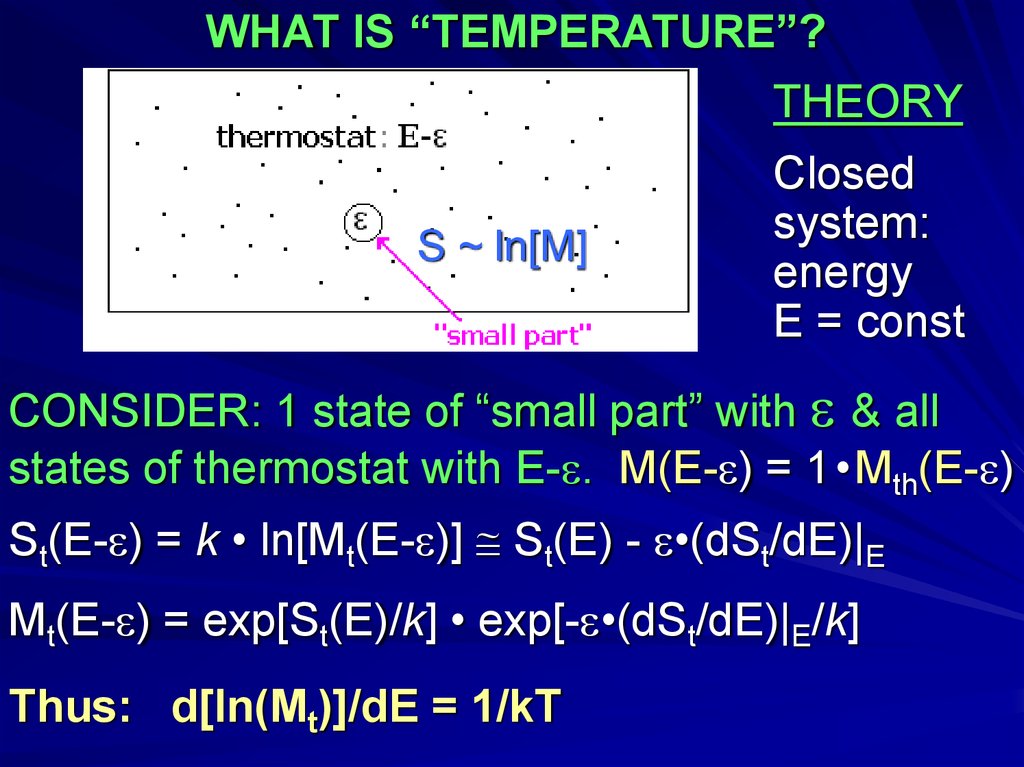
WHAT IS “TEMPERATURE”?THEORY
S ~ ln[M]
Closed
system:
energy
E = const
CONSIDER: 1 state of “small part” with & all
states of thermostat with E- . M(E- ) = 1•Mth(E- )
St(E- ) = k • ln[Mt(E- )] St(E) - •(dSt/dE)|E
Mt(E- ) = exp[St(E)/k] • exp[- •(dSt/dE)|E/k]
Thus: d[ln(Mt)]/dE = 1/kT
45.
as well:Protein structure is stable,
if its free energy is below some threshold
For example:
below that of completely unfolded chain;
or:
below that of any other globular structure
46.
More stable detail –more random sequences
Less stable detail –
less random sequences
What's good for protein’s
detail is good for the whole
protein structure
“What's good for General
Motors is good for America”
(a famous misquote of
Charles Erwin Wilson)
47.
“Multitude principle”for physical selection of folds
of globular proteins (now: “designability”):
the more sequences fit the given
architecture without destroying its stability,
the higher the occurrence of this
architecture in natural proteins.
48.
RATIONAL STRUCTURAL CLASSIFICATION OF PROTEINSGlobular
domains
C
A
T
H
S
C
O
P
49.
- Structures of water-soluble globular proteins- Physical selection of protein structures: min. of defects!
- Rational structural classification of proteins

































 physics
physics








