Similar presentations:
Complex analysis of metabolic status, intracellular pH, viscosity and cytoskeleton of human
1.
Complex analysis of metabolic status, intracellularpH, viscosity and cytoskeleton of human
mesenchymal stem cells during differentiation by
fluorescent microscopy and FLIM
A.V. Meleshina1, V.V. Dudenkova1,2, A.S. Bystrova1,2, E.V. Zagaynova1,2
1Nizhny
Novgorod State Medical Academy, Nizhny Novgorod, Russia
2Nizhny Novgorod State University, Nizhny Novgorod, Russia
2.
Functional-structural changes of MSCs during differentiationMetabolism
pH
Stem cells
High pHi=7.5-7.8
Mesenchymal
Stem
Cells
Viscosity
Viscoelastic
characteristics
Differentiation
potential
Differentiated cells
pHi=7.0-7.4
Cytoskeleton
Stem cells
Differentiated cells
fibroblast-like, spindle shape,
long, thin stress fibers
cuboidal shape, crisscrossed pattern
of actin cytoskeleton,
thick stress fibers
3.
Effective control of MSCs differentiation - great challengeComplex analysis is required!!!
4.
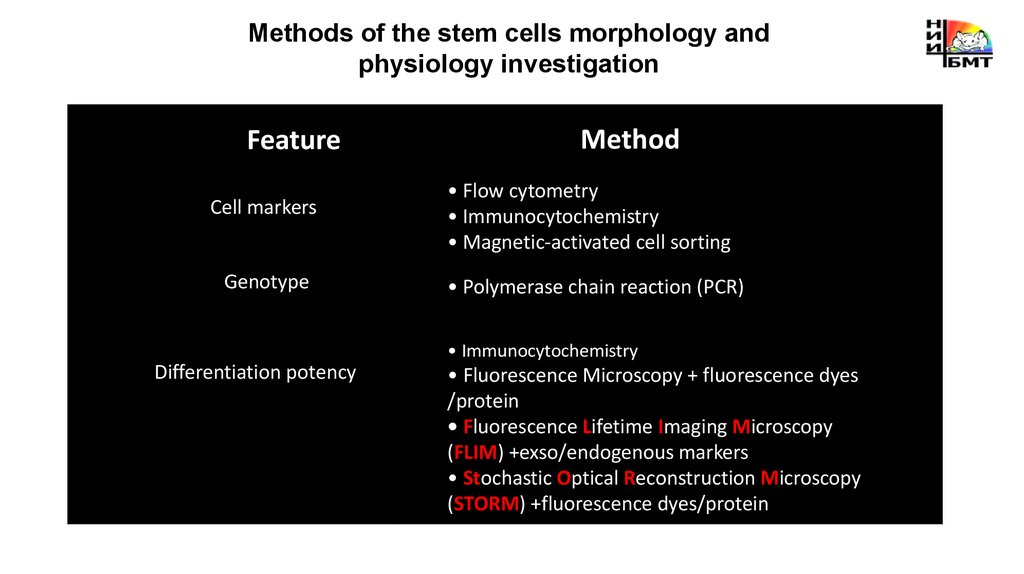
Methods of the stem cells morphology andphysiology investigation
Feature
Cell markers
Genotype
Method
• Flow cytometry
• Immunocytochemistry
• Magnetic-activated cell sorting
• Polymerase chain reaction (PCR)
• Immunocytochemistry
Differentiation potency
• Fluorescence Microscopy + fluorescence dyes
/protein
• Fluorescence Lifetime Imaging Microscopy
(FLIM) +exso/endogenous markers
• Stochastic Optical Reconstruction Microscopy
(STORM) +fluorescence dyes/protein
5.
Outline of the experiment• MSCs – human mesenchymal stem cells bone marrow
differentiated cells
0 day
7 day
14 day
21 day
chondrogenic
differentiation
Metabolism: fluorescence microscopy
pH: fluorescence microscopy and
SypHer–2
and FLIM of NAD(P)H and FAD
• YFP, monomer
• two peaks of fluorescence
excitation
(420 nm and 500 nm), peak
emission 516 nm
• at alkaline pH values, the
excitation peak at 420 nm
decreases, and at 500 nm increases,
while for acidic - on the contrary
redox ratio FAD/NAD(P)H
Lifetimes
Nicotinamide adenine dinucleotide, NADH:
excitation - 750 nm ,detection - 455-500
nm
Flavine adenine dinucleotide, FAD:
excitation - 900 nm , detection – 500-550
nm
I, a.u.
LSM 710 laser scanning confocal
microscope (Carl Zeiss, Germany)
FLIM system based on
Simple Tau 152 TCSPC system
(Becker & Hickl GmbH)
λ, nm
6.
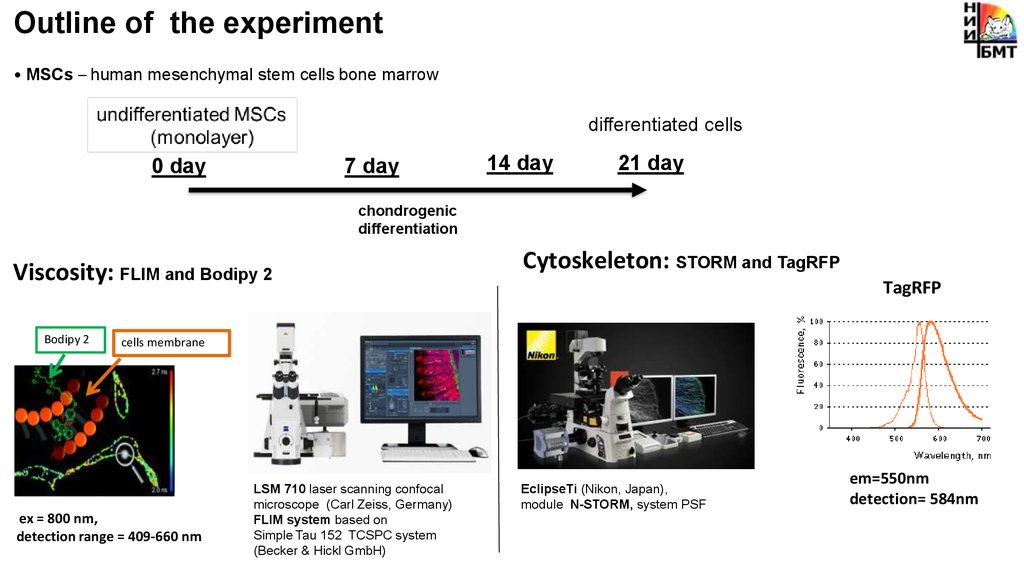
Outline of the experiment• MSCs – human mesenchymal stem cells bone marrow
differentiated cells
0 day
7 day
14 day
21 day
chondrogenic
differentiation
Viscosity: FLIM and Bodipy 2
Bodipy 2
Cytoskeleton: STORM and TagRFP
TagRFP
cells membrane
ex = 800 nm,
detection range = 409-660 nm
LSM 710 laser scanning confocal
microscope (Carl Zeiss, Germany)
FLIM system based on
Simple Tau 152 TCSPC system
(Becker & Hickl GmbH)
EclipseTi (Nikon, Japan),
module N-STORM, system PSF
em=550nm
detection= 584nm
7.
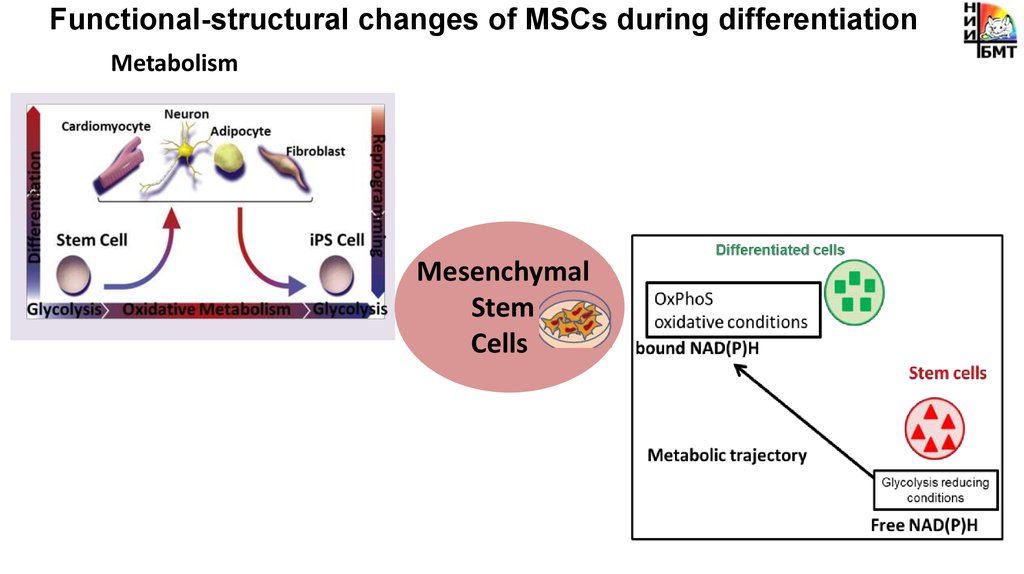
Functional-structural changes of MSCs during differentiationMetabolism
Mesenchymal
Stem
Cells
8.
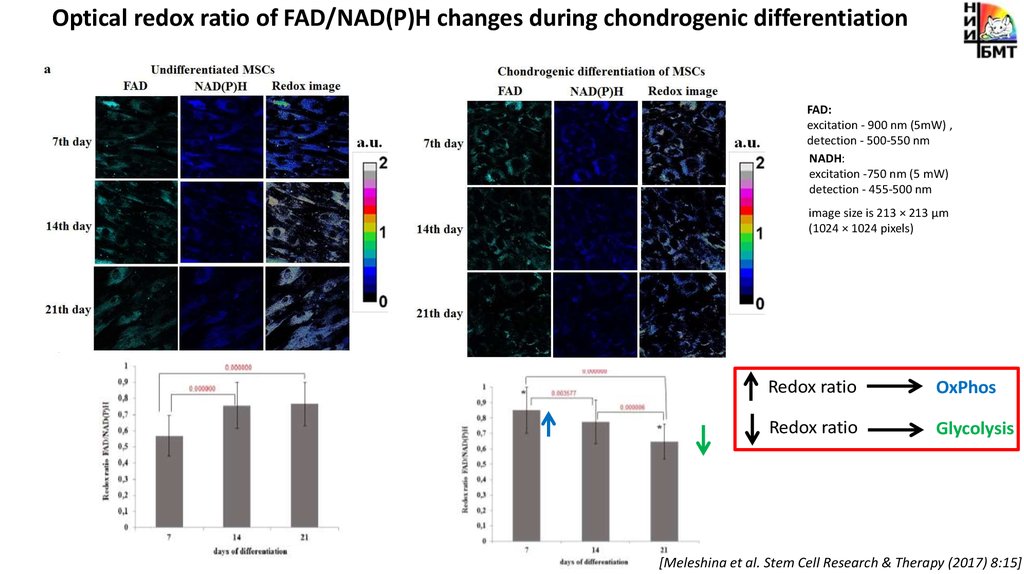
Optical redox ratio of FAD/NAD(P)H changes during chondrogenic differentiationFAD:
excitation - 900 nm (5mW) ,
detection - 500-550 nm
NADH:
excitation -750 nm (5 mW)
detection - 455-500 nm
image size is 213 × 213 μm
(1024 × 1024 pixels)
Redox ratio
OxPhos
Redox ratio
Glycolysis
[Meleshina et al. Stem Cell Research & Therapy (2017) 8:15]
9.
Dynamic of bound NAD(P)H in MSCs during chondrogenic differentiationPseudocolor-coded FLIM images of the free (t1) and protein-bound (t2) forms of
NAD(P)H.
For NAD(P)H: excitation - 750 nm, detection - 455–500 nm. Field of view
213*213μm (512*512 pixels)
[Meleshina et al. Stem Cell Research & Therapy (2017) 8:15]
10.
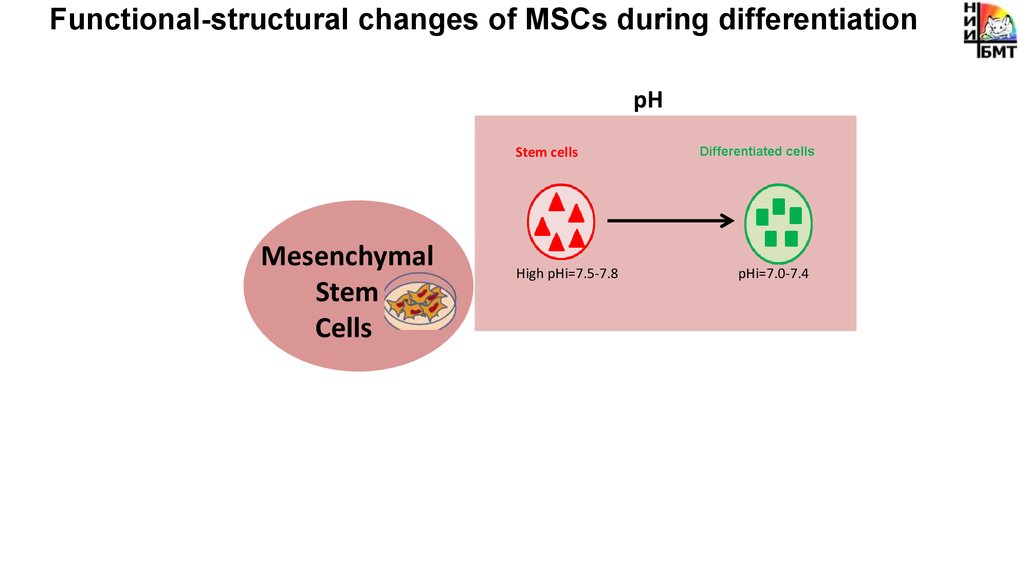
Functional-structural changes of MSCs during differentiationpH
Stem cells
Mesenchymal
Stem
Cells
High pHi=7.5-7.8
Differentiated cells
pHi=7.0-7.4
11.
Intracellular pH analysis in MSCs during differentiationby fluorescence microscopy and SypHer–2
Остеогенно дифференцированные МСК 21 день дифференцировки
Остеогенно дифференцированные
день дифференцировки
интенсивность
интенсивность МСК 21 соотношения
интенсивностей
интенсивность
интенсивность
соотношения
интенсивностей
флуоресценции
I488
флуоресценции
I
флуоресценции
(I488/I405)
Fluorescence
intensity
405
Fluorescence
intensity Redox ratio
Недифференцированные
флуоресценции I488
флуоресценции I405 МСК флуоресценции (I488/I405)
I405
интенсивность
флуоресценции I405
Undifferentiated MSCs
соотношения интенсивностей
флуоресценции (I488/I405)
Хондрогенно
дифференцированные
МСК
2121
день
дифференцировки
Хондрогенно
дифференцированные
МСК
день
дифференцировки
интенсивность
интенсивность
соотношения
интенсивностей
интенсивность
интенсивность
соотношения
интенсивностей
Остеогенно дифференцированные МСК 21 день дифференцировки
флуоресценции
I
флуоресценции
I
флуоресценции
(I(I
/I/I
))
флуоресценции
I
флуоресценции
I
флуоресценции
488
405
488
405
488
405
488
405
интенсивность
интенсивность
соотношения интенсивностей
pH, a.u.
I488
интенсивность
флуоресценции I488
Chondrogenic differentiation of MSCs
флуоресценции I488
флуоресценции I405
флуоресценции (I488/I405)
days of differentiation
Рисунок 9. Флуоресцентные изображения и изображение соотношения интенсивностей
Рисунок
9. Флуоресцентные изображения и изображение соотношения интенсивностей
(I488/I405) недифференцированных МСК и МСК на 21 день остеогенной и
ex
=флуоресценции
405
nmХондрогенно
and
detection
range =МСК
500-550
nm
флуоресценции
(I488488
/I405)nm,
недифференцированных
МСК21 идень
МСК
на 21 день остеогенной и
дифференцированные
дифференцировки
хондрогенной
дифференцировок.
Возбуждение
флуоресценции
на длине волны 488 нм и
хондрогенной
дифференцировок.
Возбуждение
флуоресценции
на
длине волны 488 нм и
интенсивность
интенсивность
соотношения
405 нм, регистрация флуоресценции в диапазоне 510-560 нм. Размеринтенсивностей
изображений 213 ×
405 213
нм,флуоресценции
регистрация
флуоресценции
в
диапазоне
510-560
нм.
Размер
изображений
флуоресценции
(I488/I405)213 ×
μm (1024 × 1024I488
pixels). флуоресценции I405
213 μm (1024 × 1024 pixels).
undifferentiated MSCs
chondrogenic differentiation
bias to acidic pH values
[unpublished data]
12.
Analysis of collagen formation during chondrogenic differentiation using SHGgreen –
cell autofluorescence
red- collagen fiber
Hematoxylin
staining
Alcian blue staining
on acidic polysaccharides
SHG of collagen was excited at wavelength of 750 nm and detected in
the range 373-387 nm
the image size is 130×130 μm (512 × 512 pixels)
[Meleshina et al. Stem Cell Research & Therapy (2017) 8:15]
13.
Functional-structural changes of MSCs during differentiationMesenchymal
Stem
Cells
Viscosity
Viscoelastic
characteristics
Differentiation
potential
Cytoskeleton
Stem cells
Differentiated cells
fibroblast-like, spindle shape,
long, thin stress fibers
cuboidal shape, crisscrossed pattern
of actin cytoskeleton,
thick stress fibers
14.
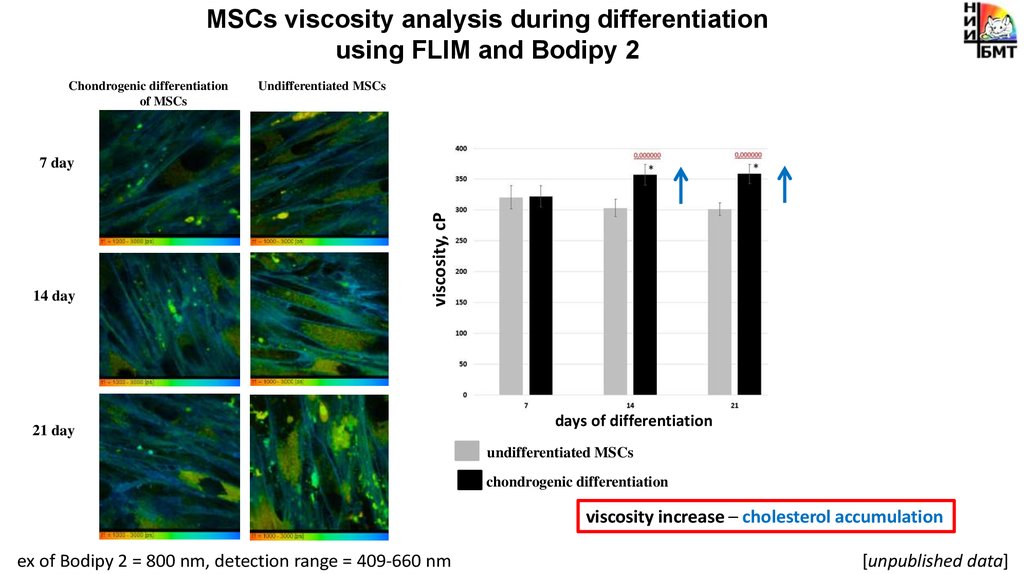
MSCs viscosity analysis during differentiationusing FLIM and Bodipy 2
Chondrogenic differentiation
of MSCs
Undifferentiated MSCs
14 day
viscosity, cP
7 day
21 day
days of differentiation
undifferentiated MSCs
chondrogenic differentiation
viscosity increase – cholesterol accumulation
ex of Bodipy 2 = 800 nm, detection range = 409-660 nm
[unpublished data]
15.
Analysis of cytoskeleton organization in MSCs during differentiationby STORM and TagRFP
Undifferentiated MSCs
7 day
14 day
21 day
Undifferentiated 7
MSCs
14
21
days of differentiation
Increase of actin fibers thickness
ex of TagRFP = 555 nm, em=584 nm
[unpublished data]
16.
take home message1. Metabolic plasticity of MSCs during chondrogenic differentiation: glycolysis –
more glycolytic state
2. Intracellular pH
bias of pH values towards a more acidic pH
3. Membrane viscosity
viscosity increase – cholesterol accumulation
4. Cytoskeleton organization
undifferentiated MSCs having a fibroblast-like morphology, the actin fibers are
represented by long, parallel fibrils extending through the cytoplasm of the cells.
Chondrocytes have increased the thickness of end parts of actin fibers. In
addition, chondrocytes have changed their orientation: actin fibrils crossed cells in
different directions
17.
AcknowledgementsThis work has been financially supported by Russian Science Foundation (grants No. 14-15-00536)
M.K. Kuimova
V.V. Dudenkova
M.V. Shirmanova
E.V. Zagaynova
A.S. Bystrova
F.A. Kulagin
N.V. Klementieva
O. Furman











 biology
biology








